| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
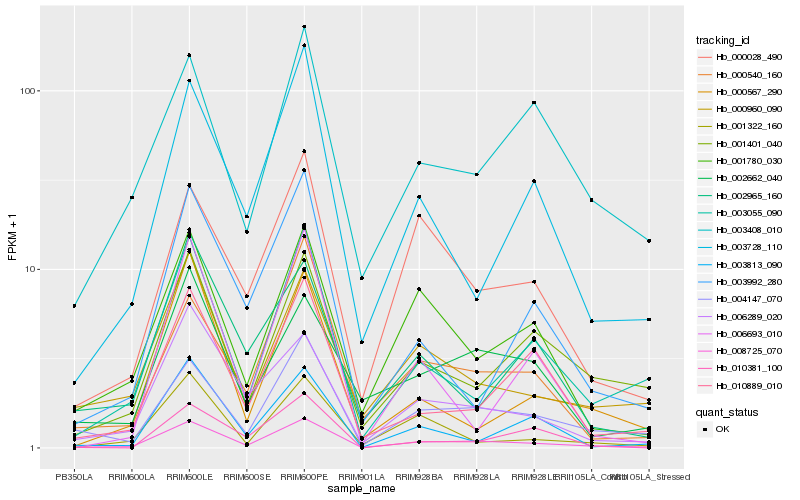
Hb_008725_070 |
0.0 |
- |
- |
Cellulose synthase A catalytic subunit 3 [UDP-forming], putative [Ricinus communis] |
| 2 |
Hb_004147_070 |
0.1527659933 |
- |
- |
PREDICTED: probable rhamnogalacturonate lyase B [Jatropha curcas] |
| 3 |
Hb_001780_030 |
0.1563919402 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 4 |
Hb_006289_020 |
0.162288441 |
- |
- |
PREDICTED: shugoshin-1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000540_160 |
0.16587489 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF6 [Jatropha curcas] |
| 6 |
Hb_000028_490 |
0.1747084531 |
- |
- |
calmodulin-like protein 6a [Populus trichocarpa] |
| 7 |
Hb_000960_090 |
0.1786083593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003728_110 |
0.1801961072 |
- |
- |
PREDICTED: calmodulin-like protein 3 [Populus euphratica] |
| 9 |
Hb_010889_010 |
0.182520592 |
- |
- |
PREDICTED: myosin-12 [Jatropha curcas] |
| 10 |
Hb_001322_160 |
0.1843179361 |
- |
- |
hypothetical protein JCGZ_03472 [Jatropha curcas] |
| 11 |
Hb_003055_090 |
0.1844611246 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 12 |
Hb_002662_040 |
0.1864984471 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 13 |
Hb_001401_040 |
0.1876503011 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 14 |
Hb_003813_090 |
0.1941302318 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 15 |
Hb_002965_160 |
0.1950944229 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 16 |
Hb_003992_280 |
0.1959959929 |
- |
- |
PREDICTED: uncharacterized protein LOC105117987 [Populus euphratica] |
| 17 |
Hb_010381_100 |
0.1983735019 |
- |
- |
Phospholipase D epsilon [Morus notabilis] |
| 18 |
Hb_006693_010 |
0.2008973009 |
- |
- |
PREDICTED: uncharacterized protein LOC105628875 [Jatropha curcas] |
| 19 |
Hb_003408_010 |
0.2010717308 |
- |
- |
fructokinase [Manihot esculenta] |
| 20 |
Hb_000567_290 |
0.2021259073 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |