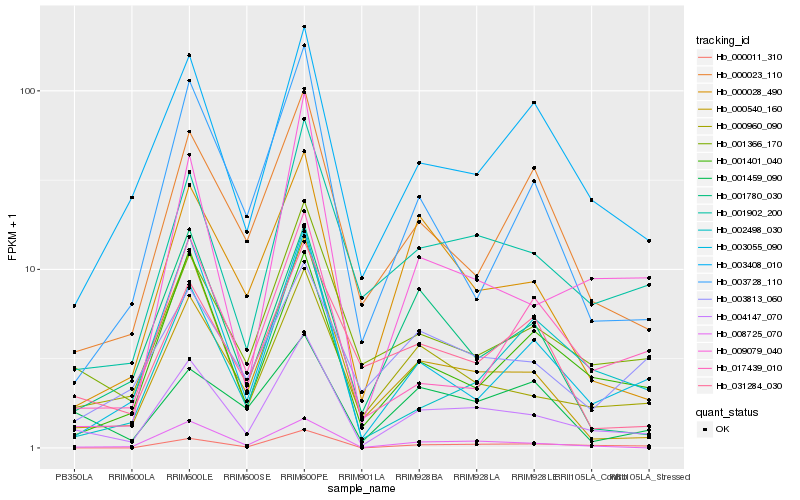
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004147_070 |
0.0 |
- |
- |
PREDICTED: probable rhamnogalacturonate lyase B [Jatropha curcas] |
| 2 |
Hb_001902_200 |
0.1411259115 |
- |
- |
PREDICTED: tubulin beta-1 chain [Eucalyptus grandis] |
| 3 |
Hb_001366_170 |
0.1464510874 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 4 |
Hb_008725_070 |
0.1527659933 |
- |
- |
Cellulose synthase A catalytic subunit 3 [UDP-forming], putative [Ricinus communis] |
| 5 |
Hb_000540_160 |
0.1624501081 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF6 [Jatropha curcas] |
| 6 |
Hb_003408_010 |
0.1669453683 |
- |
- |
fructokinase [Manihot esculenta] |
| 7 |
Hb_001401_040 |
0.1720777964 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 8 |
Hb_000011_310 |
0.1728055686 |
- |
- |
PREDICTED: uncharacterized protein LOC105631327 [Jatropha curcas] |
| 9 |
Hb_000028_490 |
0.1772072117 |
- |
- |
calmodulin-like protein 6a [Populus trichocarpa] |
| 10 |
Hb_000960_090 |
0.1807759188 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003055_090 |
0.1825476569 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 12 |
Hb_000023_110 |
0.1867279059 |
- |
- |
PREDICTED: probable methyltransferase PMT21 [Jatropha curcas] |
| 13 |
Hb_017439_010 |
0.1907044799 |
- |
- |
putative actin-97 protein [Linum usitatissimum] |
| 14 |
Hb_009079_040 |
0.1917031047 |
- |
- |
BnaA02g30070D [Brassica napus] |
| 15 |
Hb_001780_030 |
0.1917215743 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 16 |
Hb_001459_090 |
0.1929932686 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_031284_030 |
0.1959298971 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002498_030 |
0.1981136433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003813_060 |
0.1984743546 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003728_110 |
0.1988627508 |
- |
- |
PREDICTED: calmodulin-like protein 3 [Populus euphratica] |