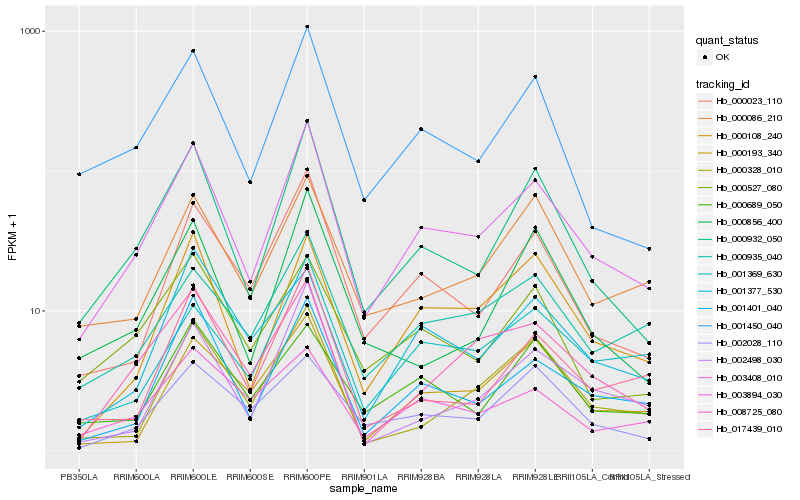
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003408_010 |
0.0 |
- |
- |
fructokinase [Manihot esculenta] |
| 2 |
Hb_000932_050 |
0.0940787448 |
- |
- |
annexin [Manihot esculenta] |
| 3 |
Hb_001450_040 |
0.1182374696 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |
| 4 |
Hb_001401_040 |
0.1191319236 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 5 |
Hb_000023_110 |
0.1194733369 |
- |
- |
PREDICTED: probable methyltransferase PMT21 [Jatropha curcas] |
| 6 |
Hb_000108_240 |
0.1297976685 |
- |
- |
hypothetical protein Csa_1G600150 [Cucumis sativus] |
| 7 |
Hb_008725_080 |
0.1393273318 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 7 [Jatropha curcas] |
| 8 |
Hb_001369_630 |
0.1420503622 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 9 |
Hb_000935_040 |
0.1452991615 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 10 |
Hb_017439_010 |
0.1460344493 |
- |
- |
putative actin-97 protein [Linum usitatissimum] |
| 11 |
Hb_000527_080 |
0.1474402396 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 12 |
Hb_000086_210 |
0.14954751 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_003894_030 |
0.150511758 |
- |
- |
plant sec1, putative [Ricinus communis] |
| 14 |
Hb_000328_010 |
0.1530822896 |
- |
- |
PREDICTED: QWRF motif-containing protein 8-like [Jatropha curcas] |
| 15 |
Hb_002498_030 |
0.1530915733 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000856_400 |
0.155123654 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 17 |
Hb_000689_050 |
0.1570222991 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000193_340 |
0.1572388109 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001377_530 |
0.1583178222 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 20 |
Hb_002028_110 |
0.1583828437 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |