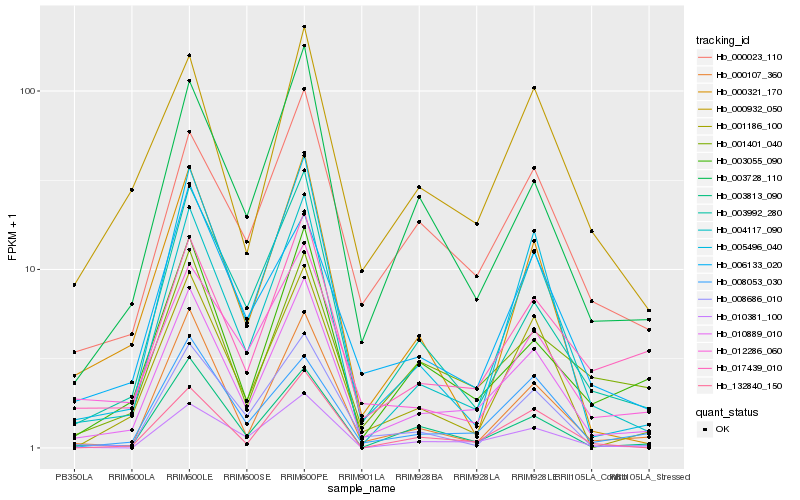
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010889_010 |
0.0 |
- |
- |
PREDICTED: myosin-12 [Jatropha curcas] |
| 2 |
Hb_004117_090 |
0.1132587403 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 70 [Jatropha curcas] |
| 3 |
Hb_010381_100 |
0.1272268376 |
- |
- |
Phospholipase D epsilon [Morus notabilis] |
| 4 |
Hb_003055_090 |
0.1280019908 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 5 |
Hb_003813_090 |
0.1291917192 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 6 |
Hb_005496_040 |
0.129619948 |
- |
- |
PREDICTED: uncharacterized protein LOC105630864 [Jatropha curcas] |
| 7 |
Hb_003728_110 |
0.1297761707 |
- |
- |
PREDICTED: calmodulin-like protein 3 [Populus euphratica] |
| 8 |
Hb_000107_360 |
0.1303652209 |
- |
- |
PREDICTED: xylem cysteine proteinase 1 [Jatropha curcas] |
| 9 |
Hb_001186_100 |
0.130992534 |
- |
- |
PREDICTED: uncharacterized protein LOC105640806 isoform X1 [Jatropha curcas] |
| 10 |
Hb_008053_030 |
0.131802744 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 11 |
Hb_003992_280 |
0.133492133 |
- |
- |
PREDICTED: uncharacterized protein LOC105117987 [Populus euphratica] |
| 12 |
Hb_017439_010 |
0.1354060137 |
- |
- |
putative actin-97 protein [Linum usitatissimum] |
| 13 |
Hb_008686_010 |
0.1407753407 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000932_050 |
0.144603141 |
- |
- |
annexin [Manihot esculenta] |
| 15 |
Hb_001401_040 |
0.1464079297 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 16 |
Hb_132840_150 |
0.1464390814 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000321_170 |
0.1467986491 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like [Jatropha curcas] |
| 18 |
Hb_006133_020 |
0.1477073049 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 19 |
Hb_000023_110 |
0.1483700394 |
- |
- |
PREDICTED: probable methyltransferase PMT21 [Jatropha curcas] |
| 20 |
Hb_012286_060 |
0.1485828061 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |