| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
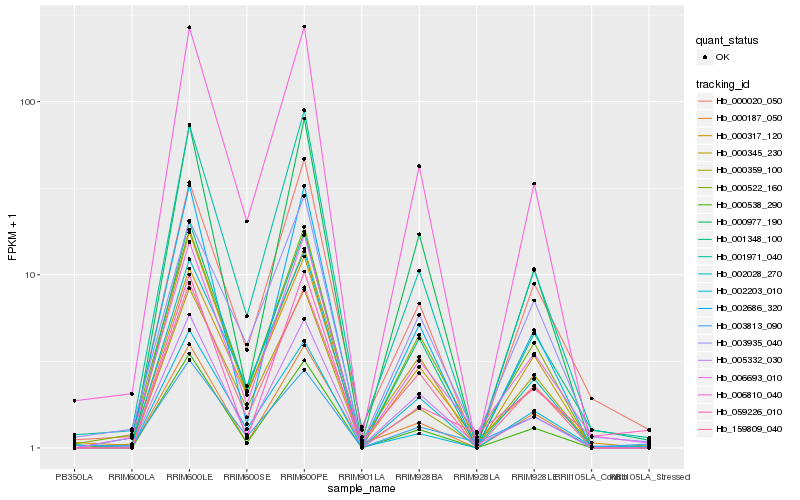
Hb_006693_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628875 [Jatropha curcas] |
| 2 |
Hb_000522_160 |
0.076679393 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 3 |
Hb_001971_040 |
0.0787861069 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 4 |
Hb_000020_050 |
0.0877896459 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 5 |
Hb_001348_100 |
0.0907734226 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |
| 6 |
Hb_006810_040 |
0.0986393907 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 7 |
Hb_005332_030 |
0.0988788582 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000359_100 |
0.1017820488 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000345_230 |
0.1086420458 |
- |
- |
- |
| 10 |
Hb_003935_040 |
0.1092705421 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP [Jatropha curcas] |
| 11 |
Hb_000538_290 |
0.1110630964 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 12 |
Hb_002686_320 |
0.1122118928 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 13 |
Hb_159809_040 |
0.1130444791 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002028_270 |
0.1131097542 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 15 |
Hb_059226_010 |
0.1139837902 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 16 |
Hb_000187_050 |
0.1153115473 |
- |
- |
PREDICTED: uncharacterized protein LOC105650800 [Jatropha curcas] |
| 17 |
Hb_003813_090 |
0.116569636 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 18 |
Hb_000977_190 |
0.1219023329 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000317_120 |
0.1222803565 |
- |
- |
PREDICTED: uncharacterized protein LOC105647000 [Jatropha curcas] |
| 20 |
Hb_002203_010 |
0.1226683602 |
- |
- |
PREDICTED: kinesin-like protein KIF18B isoform X1 [Vitis vinifera] |