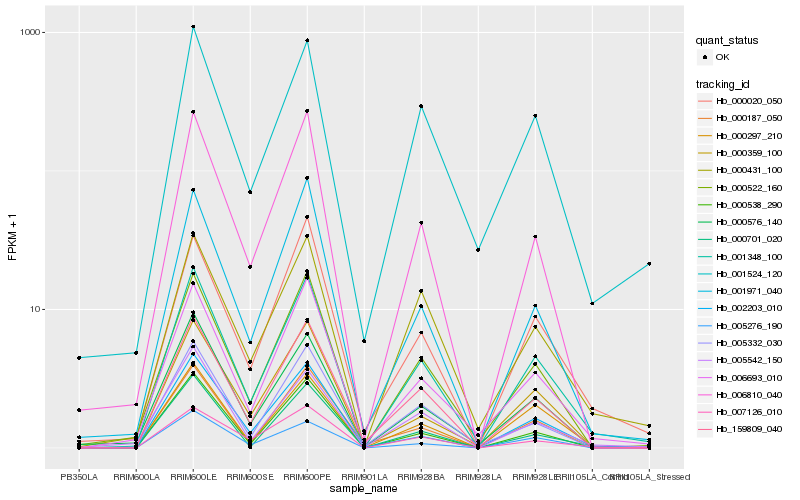
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001348_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |
| 2 |
Hb_000522_160 |
0.0894749667 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 3 |
Hb_006693_010 |
0.0907734226 |
- |
- |
PREDICTED: uncharacterized protein LOC105628875 [Jatropha curcas] |
| 4 |
Hb_159809_040 |
0.091304521 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000538_290 |
0.0951340645 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 6 |
Hb_006810_040 |
0.0956569052 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 7 |
Hb_000297_210 |
0.1012809829 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 8 |
Hb_007126_010 |
0.1016958201 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 9 |
Hb_000359_100 |
0.1034799249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000020_050 |
0.104169807 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 11 |
Hb_002203_010 |
0.1059263862 |
- |
- |
PREDICTED: kinesin-like protein KIF18B isoform X1 [Vitis vinifera] |
| 12 |
Hb_001971_040 |
0.1072118657 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 13 |
Hb_005542_150 |
0.1082690382 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 14 |
Hb_001524_120 |
0.1097950643 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 15 |
Hb_000431_100 |
0.1108579111 |
- |
- |
ribonucleoside-diphosphate reductase small chain, putative [Ricinus communis] |
| 16 |
Hb_005276_190 |
0.1120558052 |
- |
- |
PREDICTED: uncharacterized protein LOC105650003 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000576_140 |
0.1129981494 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 18 |
Hb_005332_030 |
0.1136482134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000187_050 |
0.1136489003 |
- |
- |
PREDICTED: uncharacterized protein LOC105650800 [Jatropha curcas] |
| 20 |
Hb_000701_020 |
0.1142120164 |
- |
- |
PREDICTED: protein ENDOSPERM DEFECTIVE 1-like isoform X1 [Jatropha curcas] |