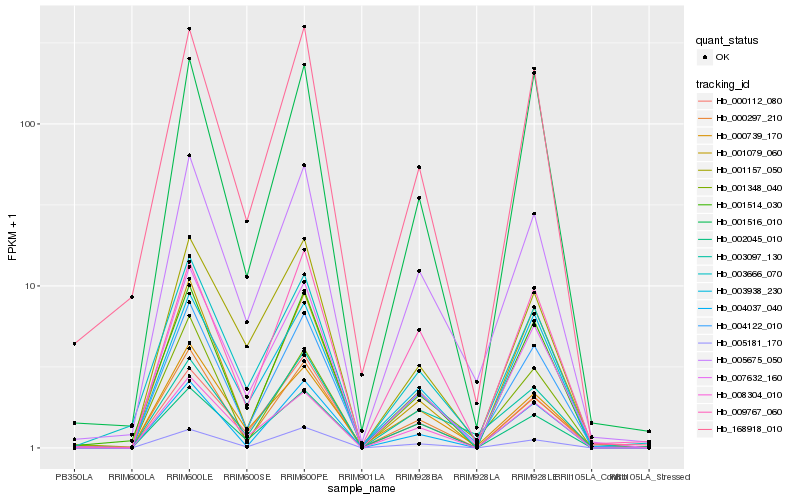
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004122_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630923 [Jatropha curcas] |
| 2 |
Hb_005675_050 |
0.0626286124 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 3 |
Hb_000297_210 |
0.0820815603 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 4 |
Hb_001514_030 |
0.0854152454 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 5 |
Hb_007632_160 |
0.0863104522 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_168918_010 |
0.091547802 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 7 |
Hb_005181_170 |
0.0974786572 |
- |
- |
PREDICTED: aldehyde dehydrogenase 22A1 [Jatropha curcas] |
| 8 |
Hb_000739_170 |
0.0993257803 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 9 |
Hb_009767_060 |
0.0995743568 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 10 |
Hb_001079_060 |
0.1001816918 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_008304_010 |
0.1009190223 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 12 |
Hb_003938_230 |
0.1025419761 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 13 |
Hb_003666_070 |
0.1028902694 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |
| 14 |
Hb_004037_040 |
0.1040990522 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase [Jatropha curcas] |
| 15 |
Hb_001516_010 |
0.108822958 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 16 |
Hb_001348_040 |
0.1097150685 |
- |
- |
hypothetical protein VITISV_006651 [Vitis vinifera] |
| 17 |
Hb_002045_010 |
0.1105954817 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 17 [Jatropha curcas] |
| 18 |
Hb_003097_130 |
0.1157651062 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000112_080 |
0.1159859058 |
- |
- |
PREDICTED: glutamate receptor 2.1-like [Populus euphratica] |
| 20 |
Hb_001157_050 |
0.1195335918 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |