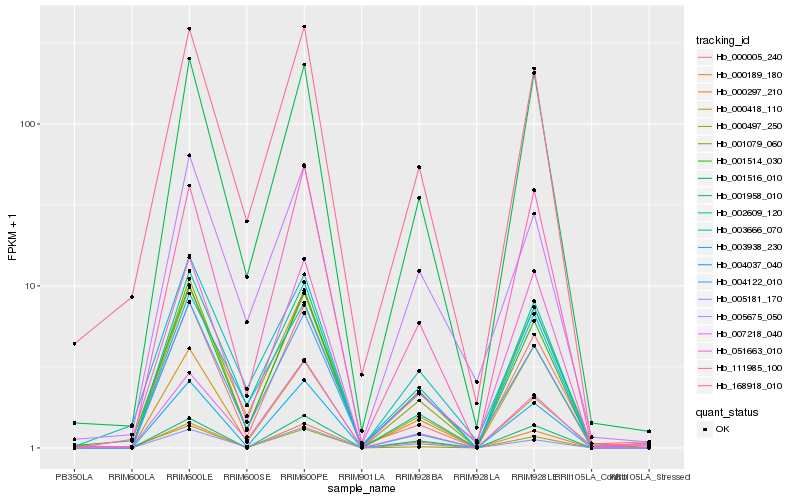
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001514_030 |
0.0 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 2 |
Hb_168918_010 |
0.0513426014 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 3 |
Hb_004037_040 |
0.0713377366 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase [Jatropha curcas] |
| 4 |
Hb_001079_060 |
0.0803740992 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001516_010 |
0.0832563375 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 6 |
Hb_004122_010 |
0.0854152454 |
- |
- |
PREDICTED: uncharacterized protein LOC105630923 [Jatropha curcas] |
| 7 |
Hb_007218_040 |
0.0863885217 |
- |
- |
Leucine-rich repeat transmembrane protein kinase protein, putative [Theobroma cacao] |
| 8 |
Hb_051663_010 |
0.0957976671 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 9 |
Hb_003938_230 |
0.1038300301 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 10 |
Hb_005181_170 |
0.1042874945 |
- |
- |
PREDICTED: aldehyde dehydrogenase 22A1 [Jatropha curcas] |
| 11 |
Hb_000189_180 |
0.1053685812 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 12 |
Hb_003666_070 |
0.1056000219 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |
| 13 |
Hb_002609_120 |
0.1067877739 |
- |
- |
hypothetical protein POPTR_0001s25660g [Populus trichocarpa] |
| 14 |
Hb_111985_100 |
0.1079762901 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 15 |
Hb_000497_250 |
0.1093211481 |
- |
- |
PREDICTED: potassium transporter 5 [Jatropha curcas] |
| 16 |
Hb_001958_010 |
0.1098397366 |
- |
- |
hypothetical protein CICLE_v100078532mg, partial [Citrus clementina] |
| 17 |
Hb_000418_110 |
0.1100862999 |
- |
- |
PREDICTED: protein kinase PINOID [Jatropha curcas] |
| 18 |
Hb_000005_240 |
0.1106887551 |
- |
- |
Shugoshin-1, putative [Ricinus communis] |
| 19 |
Hb_000297_210 |
0.1109146944 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 20 |
Hb_005675_050 |
0.1116959053 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |