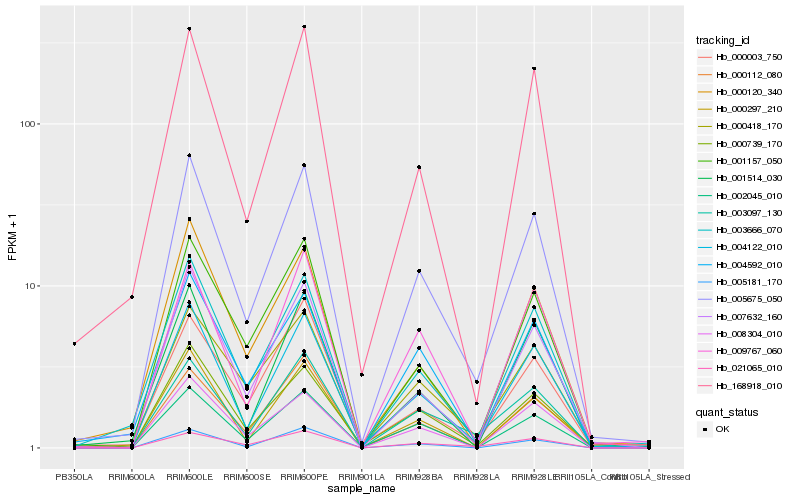
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005675_050 |
0.0 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 2 |
Hb_004122_010 |
0.0626286124 |
- |
- |
PREDICTED: uncharacterized protein LOC105630923 [Jatropha curcas] |
| 3 |
Hb_003666_070 |
0.0800614576 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |
| 4 |
Hb_007632_160 |
0.0866677588 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000739_170 |
0.0937779811 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 6 |
Hb_003097_130 |
0.0942196306 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000418_170 |
0.0956657849 |
- |
- |
PREDICTED: ABC transporter C family member 12-like isoform X4 [Populus euphratica] |
| 8 |
Hb_001157_050 |
0.0983896358 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 9 |
Hb_168918_010 |
0.1004378853 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 10 |
Hb_000297_210 |
0.1041587238 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 11 |
Hb_008304_010 |
0.106829024 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 12 |
Hb_005181_170 |
0.1076316574 |
- |
- |
PREDICTED: aldehyde dehydrogenase 22A1 [Jatropha curcas] |
| 13 |
Hb_004592_010 |
0.1076875279 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b-like [Jatropha curcas] |
| 14 |
Hb_021065_010 |
0.1077236886 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like isoform X2 [Gossypium raimondii] |
| 15 |
Hb_000003_750 |
0.1090095687 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 16 |
Hb_009767_060 |
0.1092985533 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 17 |
Hb_002045_010 |
0.1095970439 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 17 [Jatropha curcas] |
| 18 |
Hb_000120_340 |
0.1096060244 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 19 |
Hb_000112_080 |
0.1114629879 |
- |
- |
PREDICTED: glutamate receptor 2.1-like [Populus euphratica] |
| 20 |
Hb_001514_030 |
0.1116959053 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |