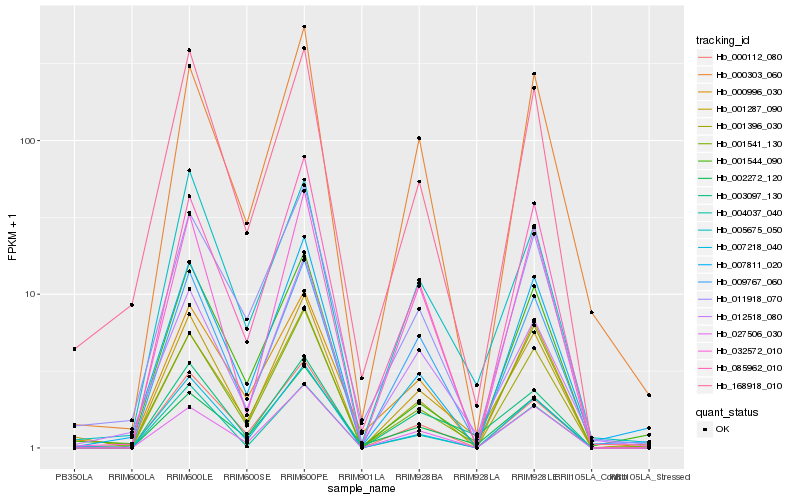
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001287_090 |
0.0 |
- |
- |
Endoglucanase 4 [Aegilops tauschii] |
| 2 |
Hb_009767_060 |
0.1013984571 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 3 |
Hb_001396_030 |
0.1031253651 |
- |
- |
hypothetical protein POPTR_0001s13520g [Populus trichocarpa] |
| 4 |
Hb_001541_130 |
0.1073428855 |
- |
- |
GDSL esterase/lipase [Morus notabilis] |
| 5 |
Hb_168918_010 |
0.1114994404 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 6 |
Hb_085962_010 |
0.1124083563 |
- |
- |
hypothetical protein POPTR_0017s11380g [Populus trichocarpa] |
| 7 |
Hb_007811_020 |
0.1133136307 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 8 |
Hb_000303_060 |
0.114301567 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_011918_070 |
0.1146692864 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 10 |
Hb_004037_040 |
0.1149619294 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase [Jatropha curcas] |
| 11 |
Hb_002272_120 |
0.1150775409 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 12 |
Hb_003097_130 |
0.1158359811 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_027506_030 |
0.1167153241 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 14 |
Hb_007218_040 |
0.1175966429 |
- |
- |
Leucine-rich repeat transmembrane protein kinase protein, putative [Theobroma cacao] |
| 15 |
Hb_000112_080 |
0.1182506916 |
- |
- |
PREDICTED: glutamate receptor 2.1-like [Populus euphratica] |
| 16 |
Hb_000996_030 |
0.1186578836 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g63430 isoform X1 [Jatropha curcas] |
| 17 |
Hb_032572_010 |
0.1190547519 |
- |
- |
PREDICTED: phospholipase A2-alpha [Jatropha curcas] |
| 18 |
Hb_001544_090 |
0.1207569673 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 19 |
Hb_012518_080 |
0.1208924217 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like [Jatropha curcas] |
| 20 |
Hb_005675_050 |
0.1210775213 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |