| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
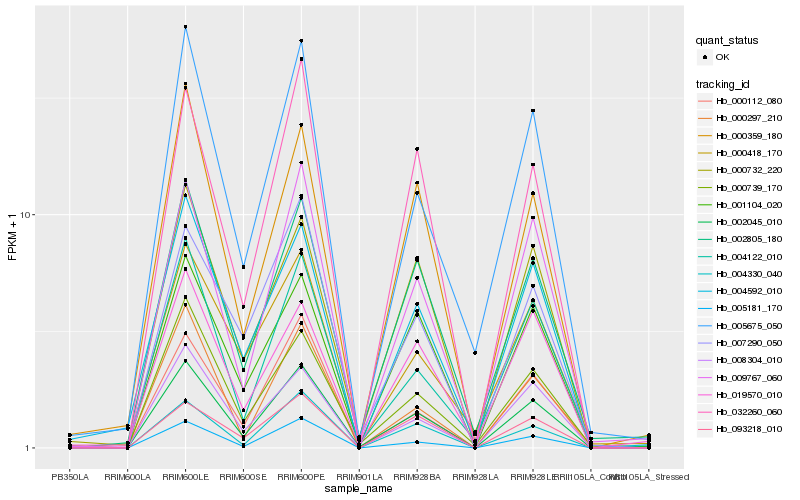
Hb_002045_010 |
0.0 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 17 [Jatropha curcas] |
| 2 |
Hb_002805_180 |
0.0851052937 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_009767_060 |
0.0895783354 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 4 |
Hb_000359_180 |
0.1041918003 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005181_170 |
0.1043945791 |
- |
- |
PREDICTED: aldehyde dehydrogenase 22A1 [Jatropha curcas] |
| 6 |
Hb_000739_170 |
0.1050474334 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 7 |
Hb_004330_040 |
0.1078885707 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_032260_060 |
0.1081539434 |
transcription factor |
TF Family: MYB-related |
Homeodomain-like superfamily protein [Theobroma cacao] |
| 9 |
Hb_000297_210 |
0.1090717098 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 10 |
Hb_005675_050 |
0.1095970439 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 11 |
Hb_000112_080 |
0.1096812462 |
- |
- |
PREDICTED: glutamate receptor 2.1-like [Populus euphratica] |
| 12 |
Hb_004122_010 |
0.1105954817 |
- |
- |
PREDICTED: uncharacterized protein LOC105630923 [Jatropha curcas] |
| 13 |
Hb_001104_020 |
0.1117833169 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000732_220 |
0.1125623587 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 15 |
Hb_004592_010 |
0.113079487 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b-like [Jatropha curcas] |
| 16 |
Hb_019570_010 |
0.1163358357 |
- |
- |
lyase, putative [Ricinus communis] |
| 17 |
Hb_008304_010 |
0.1163445032 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 18 |
Hb_007290_050 |
0.1200912469 |
- |
- |
PREDICTED: uncharacterized protein LOC105629472 isoform X1 [Jatropha curcas] |
| 19 |
Hb_093218_010 |
0.1251505967 |
- |
- |
hypothetical protein POPTR_0001s05220g [Populus trichocarpa] |
| 20 |
Hb_000418_170 |
0.1253810839 |
- |
- |
PREDICTED: ABC transporter C family member 12-like isoform X4 [Populus euphratica] |