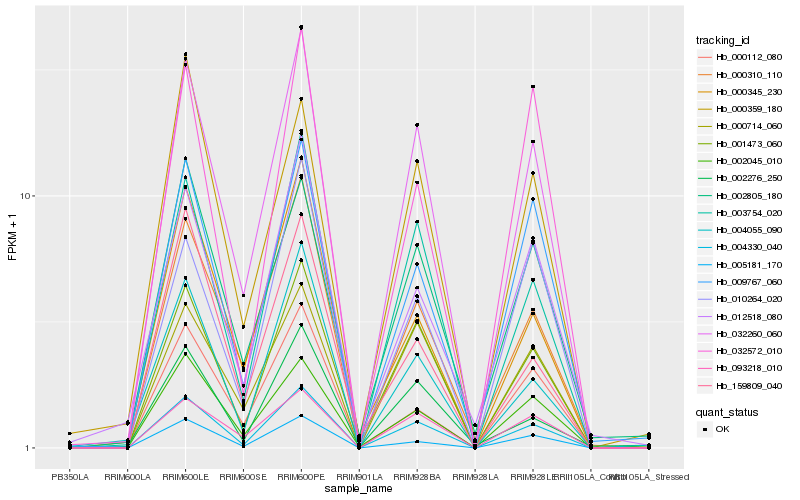
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004330_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_032260_060 |
0.0416453697 |
transcription factor |
TF Family: MYB-related |
Homeodomain-like superfamily protein [Theobroma cacao] |
| 3 |
Hb_001473_060 |
0.0659324221 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 8 [Jatropha curcas] |
| 4 |
Hb_003754_020 |
0.0713017216 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_005181_170 |
0.1025348162 |
- |
- |
PREDICTED: aldehyde dehydrogenase 22A1 [Jatropha curcas] |
| 6 |
Hb_000310_110 |
0.1037992608 |
- |
- |
PREDICTED: probable trehalose-phosphate phosphatase F isoform X1 [Jatropha curcas] |
| 7 |
Hb_002045_010 |
0.1078885707 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 17 [Jatropha curcas] |
| 8 |
Hb_009767_060 |
0.1127747662 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 9 |
Hb_093218_010 |
0.1135553867 |
- |
- |
hypothetical protein POPTR_0001s05220g [Populus trichocarpa] |
| 10 |
Hb_000714_060 |
0.1143260407 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 1-like [Populus euphratica] |
| 11 |
Hb_004055_090 |
0.1144197158 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 12 |
Hb_000112_080 |
0.1257059083 |
- |
- |
PREDICTED: glutamate receptor 2.1-like [Populus euphratica] |
| 13 |
Hb_002805_180 |
0.1296895821 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000345_230 |
0.1305543695 |
- |
- |
- |
| 15 |
Hb_002276_250 |
0.1317797439 |
- |
- |
PREDICTED: WD repeat and HMG-box DNA-binding protein 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_032572_010 |
0.1331720852 |
- |
- |
PREDICTED: phospholipase A2-alpha [Jatropha curcas] |
| 17 |
Hb_010264_020 |
0.133744735 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000359_180 |
0.1358981758 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_012518_080 |
0.1371385674 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like [Jatropha curcas] |
| 20 |
Hb_159809_040 |
0.1413462794 |
- |
- |
conserved hypothetical protein [Ricinus communis] |