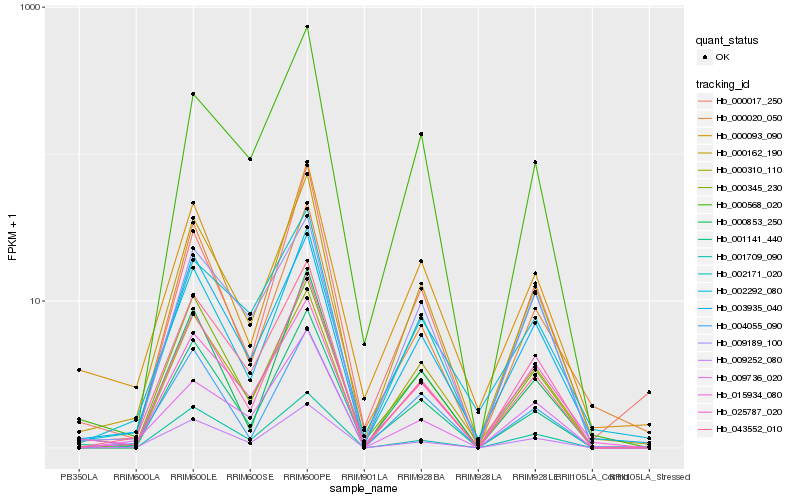
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000162_190 |
0.0 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 2 |
Hb_003935_040 |
0.0759100258 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP [Jatropha curcas] |
| 3 |
Hb_025787_020 |
0.0791731698 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 4 |
Hb_009252_080 |
0.0823968995 |
- |
- |
Polygalacturonase-1 non-catalytic subunit beta precursor, putative [Ricinus communis] |
| 5 |
Hb_001141_440 |
0.0867471757 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 6 |
Hb_043552_010 |
0.091391723 |
- |
- |
PREDICTED: pectinesterase/pectinesterase inhibitor PPE8B isoform X1 [Jatropha curcas] |
| 7 |
Hb_000310_110 |
0.0947021651 |
- |
- |
PREDICTED: probable trehalose-phosphate phosphatase F isoform X1 [Jatropha curcas] |
| 8 |
Hb_015934_080 |
0.0971813964 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 9 |
Hb_000345_230 |
0.0978841975 |
- |
- |
- |
| 10 |
Hb_002292_080 |
0.0989310508 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 11 |
Hb_000853_250 |
0.1011972579 |
- |
- |
Cellulose synthase A catalytic subunit 3 [UDP-forming], putative [Ricinus communis] |
| 12 |
Hb_000093_090 |
0.1026667019 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |
| 13 |
Hb_009189_100 |
0.1026903061 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 14 |
Hb_000568_020 |
0.1047810395 |
- |
- |
PREDICTED: probable aquaporin PIP2-5 [Jatropha curcas] |
| 15 |
Hb_002171_020 |
0.1050994615 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 16 |
Hb_000017_250 |
0.1101878463 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 17 |
Hb_004055_090 |
0.1122274646 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 18 |
Hb_001709_090 |
0.1122623985 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 19 |
Hb_009736_020 |
0.1133712311 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG13 homolog [Jatropha curcas] |
| 20 |
Hb_000020_050 |
0.1150326587 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |