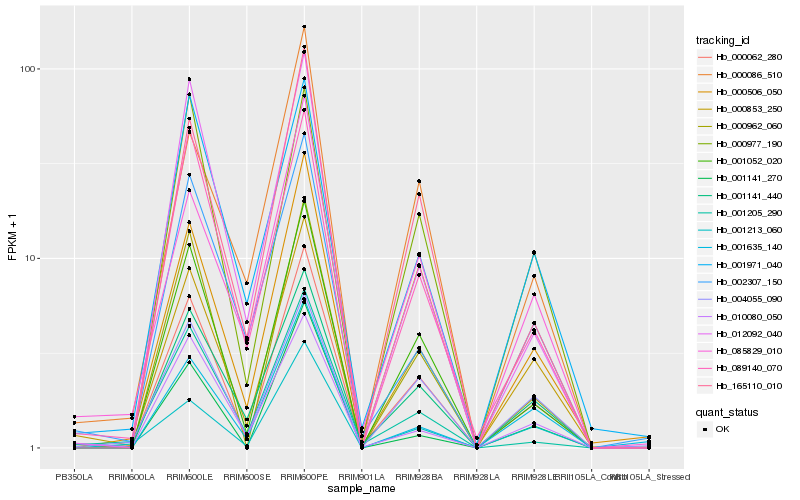
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_280 |
0.0 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |
| 2 |
Hb_001052_020 |
0.0682971345 |
- |
- |
hypothetical protein JCGZ_26487 [Jatropha curcas] |
| 3 |
Hb_000853_250 |
0.0871060382 |
- |
- |
Cellulose synthase A catalytic subunit 3 [UDP-forming], putative [Ricinus communis] |
| 4 |
Hb_165110_010 |
0.0917878556 |
- |
- |
PREDICTED: kunitz-type elastase inhibitor BrEI-like [Jatropha curcas] |
| 5 |
Hb_001141_440 |
0.0971132754 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 6 |
Hb_000962_060 |
0.0998181344 |
- |
- |
PREDICTED: uncharacterized protein LOC105646517 [Jatropha curcas] |
| 7 |
Hb_000506_050 |
0.103370111 |
- |
- |
PREDICTED: thymidine kinase [Jatropha curcas] |
| 8 |
Hb_089140_070 |
0.1222194493 |
- |
- |
PREDICTED: uncharacterized protein LOC105637961 [Jatropha curcas] |
| 9 |
Hb_085829_010 |
0.1230528107 |
- |
- |
PREDICTED: homeobox protein 2-like [Jatropha curcas] |
| 10 |
Hb_004055_090 |
0.1272322116 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 11 |
Hb_000977_190 |
0.1276288213 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001213_060 |
0.1285332246 |
- |
- |
PREDICTED: uncharacterized protein LOC105646662 [Jatropha curcas] |
| 13 |
Hb_001205_290 |
0.1300445565 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP7-like [Jatropha curcas] |
| 14 |
Hb_001971_040 |
0.1334728438 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 15 |
Hb_010080_050 |
0.1348346787 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 16 |
Hb_012092_040 |
0.1364729322 |
- |
- |
PREDICTED: expansin-A1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001635_140 |
0.1372361631 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_002307_150 |
0.1374287473 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA4-like [Jatropha curcas] |
| 19 |
Hb_000086_510 |
0.1377993662 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_001141_270 |
0.1379484784 |
- |
- |
PREDICTED: uncharacterized protein LOC105632005 [Jatropha curcas] |