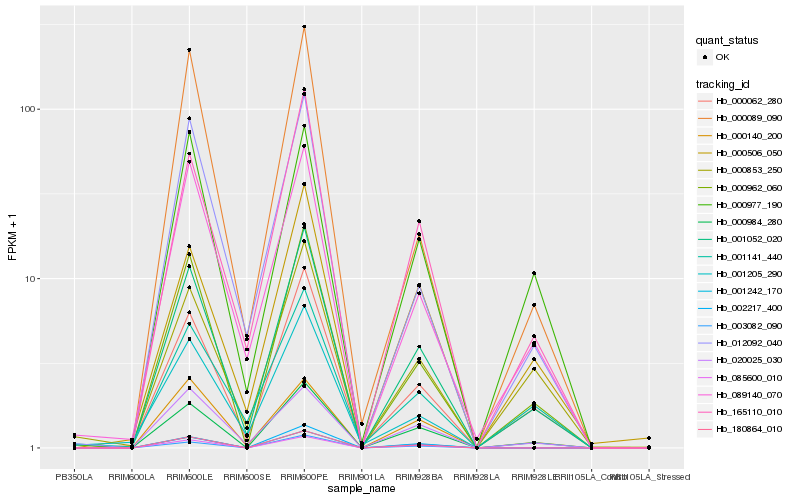
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001052_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_26487 [Jatropha curcas] |
| 2 |
Hb_000062_280 |
0.0682971345 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |
| 3 |
Hb_165110_010 |
0.0712527393 |
- |
- |
PREDICTED: kunitz-type elastase inhibitor BrEI-like [Jatropha curcas] |
| 4 |
Hb_000962_060 |
0.0774845435 |
- |
- |
PREDICTED: uncharacterized protein LOC105646517 [Jatropha curcas] |
| 5 |
Hb_089140_070 |
0.1079492335 |
- |
- |
PREDICTED: uncharacterized protein LOC105637961 [Jatropha curcas] |
| 6 |
Hb_000506_050 |
0.1152925504 |
- |
- |
PREDICTED: thymidine kinase [Jatropha curcas] |
| 7 |
Hb_012092_040 |
0.1154934114 |
- |
- |
PREDICTED: expansin-A1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_180864_010 |
0.1167050709 |
- |
- |
hypothetical protein B456_010G097000 [Gossypium raimondii] |
| 9 |
Hb_020025_030 |
0.1196215902 |
transcription factor |
TF Family: PHD |
PREDICTED: protein Jade-1 [Populus euphratica] |
| 10 |
Hb_001242_170 |
0.1202858813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000977_190 |
0.12275842 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000089_090 |
0.1229148045 |
- |
- |
hypothetical protein POPTR_0019s12670g [Populus trichocarpa] |
| 13 |
Hb_000140_200 |
0.1233955382 |
- |
- |
PREDICTED: uncharacterized protein LOC105642928 [Jatropha curcas] |
| 14 |
Hb_000984_280 |
0.1238296505 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2 [Jatropha curcas] |
| 15 |
Hb_085600_010 |
0.1249950297 |
- |
- |
PREDICTED: peroxidase 24-like [Jatropha curcas] |
| 16 |
Hb_000853_250 |
0.1250268685 |
- |
- |
Cellulose synthase A catalytic subunit 3 [UDP-forming], putative [Ricinus communis] |
| 17 |
Hb_003082_090 |
0.1255695388 |
- |
- |
hypothetical protein POPTR_0013s099451g, partial [Populus trichocarpa] |
| 18 |
Hb_001205_290 |
0.1257292327 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP7-like [Jatropha curcas] |
| 19 |
Hb_001141_440 |
0.126101176 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 20 |
Hb_002217_400 |
0.1265964068 |
- |
- |
polynucleotidyl transferase, ribonuclease H-like protein [Arabidopsis thaliana] |