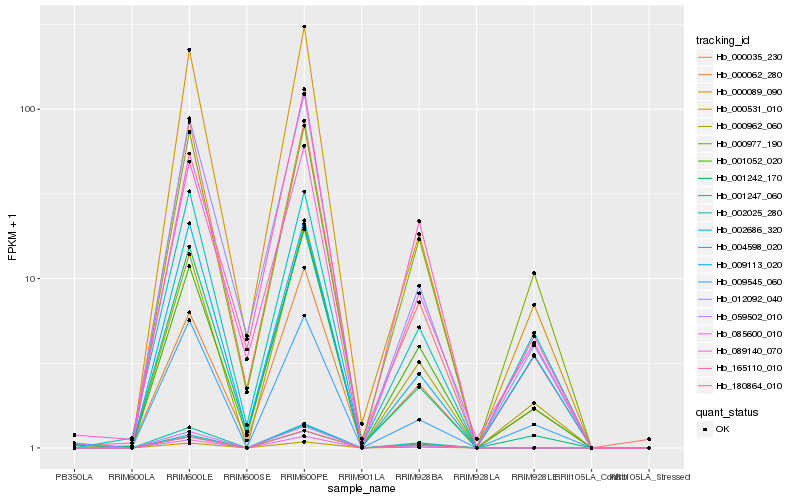
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000962_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646517 [Jatropha curcas] |
| 2 |
Hb_001052_020 |
0.0774845435 |
- |
- |
hypothetical protein JCGZ_26487 [Jatropha curcas] |
| 3 |
Hb_009545_060 |
0.087900352 |
- |
- |
PREDICTED: uncharacterized protein LOC105632005 [Jatropha curcas] |
| 4 |
Hb_000062_280 |
0.0998181344 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |
| 5 |
Hb_000089_090 |
0.1013204492 |
- |
- |
hypothetical protein POPTR_0019s12670g [Populus trichocarpa] |
| 6 |
Hb_012092_040 |
0.1104378999 |
- |
- |
PREDICTED: expansin-A1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001247_060 |
0.1113570706 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_089140_070 |
0.1129392907 |
- |
- |
PREDICTED: uncharacterized protein LOC105637961 [Jatropha curcas] |
| 9 |
Hb_059502_010 |
0.1188956119 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 10 |
Hb_000531_010 |
0.1192502386 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0018s14830g [Populus trichocarpa] |
| 11 |
Hb_180864_010 |
0.1215394602 |
- |
- |
hypothetical protein B456_010G097000 [Gossypium raimondii] |
| 12 |
Hb_004598_020 |
0.1224560139 |
transcription factor |
TF Family: ERF |
Transcriptional factor TINY, putative [Ricinus communis] |
| 13 |
Hb_000035_230 |
0.1252968599 |
- |
- |
PREDICTED: early nodulin-like protein 1 [Jatropha curcas] |
| 14 |
Hb_009113_020 |
0.1259014454 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 15 |
Hb_000977_190 |
0.126167659 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002686_320 |
0.1283876888 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 17 |
Hb_001242_170 |
0.1320168631 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_085600_010 |
0.1349228495 |
- |
- |
PREDICTED: peroxidase 24-like [Jatropha curcas] |
| 19 |
Hb_165110_010 |
0.1357391528 |
- |
- |
PREDICTED: kunitz-type elastase inhibitor BrEI-like [Jatropha curcas] |
| 20 |
Hb_002025_280 |
0.135932108 |
- |
- |
PREDICTED: uncharacterized protein LOC105634201 [Jatropha curcas] |