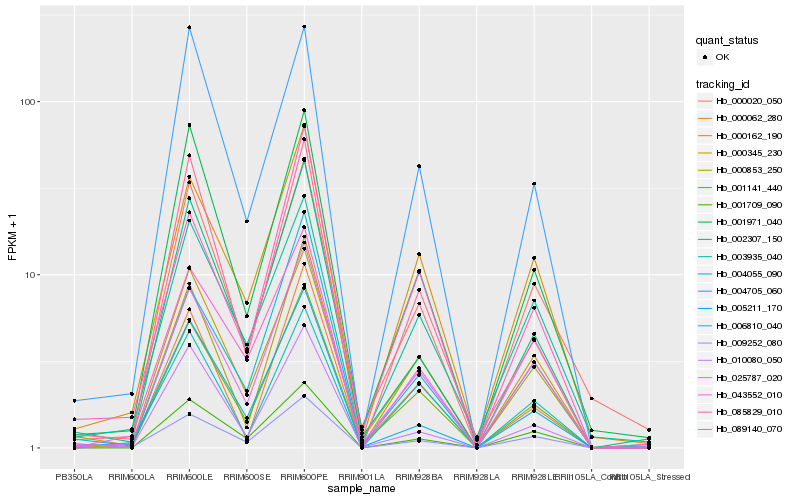
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001141_440 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 2 |
Hb_000853_250 |
0.0711807792 |
- |
- |
Cellulose synthase A catalytic subunit 3 [UDP-forming], putative [Ricinus communis] |
| 3 |
Hb_000162_190 |
0.0867471757 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 4 |
Hb_000345_230 |
0.0885585095 |
- |
- |
- |
| 5 |
Hb_001971_040 |
0.0952209288 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 6 |
Hb_000062_280 |
0.0971132754 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |
| 7 |
Hb_009252_080 |
0.0998469243 |
- |
- |
Polygalacturonase-1 non-catalytic subunit beta precursor, putative [Ricinus communis] |
| 8 |
Hb_002307_150 |
0.1010663613 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA4-like [Jatropha curcas] |
| 9 |
Hb_043552_010 |
0.1034669386 |
- |
- |
PREDICTED: pectinesterase/pectinesterase inhibitor PPE8B isoform X1 [Jatropha curcas] |
| 10 |
Hb_025787_020 |
0.1081372398 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 11 |
Hb_010080_050 |
0.1089272216 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 12 |
Hb_005211_170 |
0.1089788079 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 13 |
Hb_000020_050 |
0.1091880095 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 14 |
Hb_004055_090 |
0.1092206725 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 15 |
Hb_089140_070 |
0.1104253244 |
- |
- |
PREDICTED: uncharacterized protein LOC105637961 [Jatropha curcas] |
| 16 |
Hb_001709_090 |
0.110601771 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 17 |
Hb_004705_060 |
0.1132444015 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 18 |
Hb_006810_040 |
0.1143609008 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 19 |
Hb_085829_010 |
0.1148389118 |
- |
- |
PREDICTED: homeobox protein 2-like [Jatropha curcas] |
| 20 |
Hb_003935_040 |
0.1184867361 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP [Jatropha curcas] |