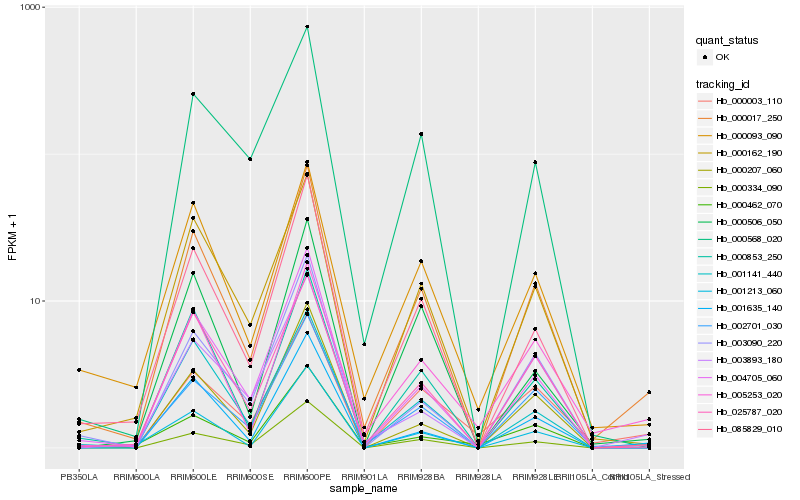
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000017_250 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 2 |
Hb_002701_030 |
0.0863431445 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 3 |
Hb_004705_060 |
0.0971079659 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 4 |
Hb_085829_010 |
0.0992996879 |
- |
- |
PREDICTED: homeobox protein 2-like [Jatropha curcas] |
| 5 |
Hb_000853_250 |
0.1057664795 |
- |
- |
Cellulose synthase A catalytic subunit 3 [UDP-forming], putative [Ricinus communis] |
| 6 |
Hb_000162_190 |
0.1101878463 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 7 |
Hb_025787_020 |
0.1109715618 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 8 |
Hb_001635_140 |
0.1123779298 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_000334_090 |
0.1199079816 |
- |
- |
hypothetical protein JCGZ_16789 [Jatropha curcas] |
| 10 |
Hb_000093_090 |
0.1256738926 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |
| 11 |
Hb_000207_060 |
0.1271243328 |
- |
- |
PREDICTED: uncharacterized protein LOC105634368 [Jatropha curcas] |
| 12 |
Hb_001141_440 |
0.1283086349 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 13 |
Hb_003893_180 |
0.1331042651 |
- |
- |
PREDICTED: uncharacterized protein LOC105648771 [Jatropha curcas] |
| 14 |
Hb_005253_020 |
0.1331353102 |
- |
- |
PREDICTED: uncharacterized protein LOC105629998 [Jatropha curcas] |
| 15 |
Hb_003090_220 |
0.1332510576 |
- |
- |
PREDICTED: copper transporter 4 [Jatropha curcas] |
| 16 |
Hb_001213_060 |
0.1341625915 |
- |
- |
PREDICTED: uncharacterized protein LOC105646662 [Jatropha curcas] |
| 17 |
Hb_000462_070 |
0.1347630346 |
- |
- |
PREDICTED: probable pectinesterase 15 [Jatropha curcas] |
| 18 |
Hb_000506_050 |
0.1369035742 |
- |
- |
PREDICTED: thymidine kinase [Jatropha curcas] |
| 19 |
Hb_000568_020 |
0.136909369 |
- |
- |
PREDICTED: probable aquaporin PIP2-5 [Jatropha curcas] |
| 20 |
Hb_000003_110 |
0.1377811694 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |