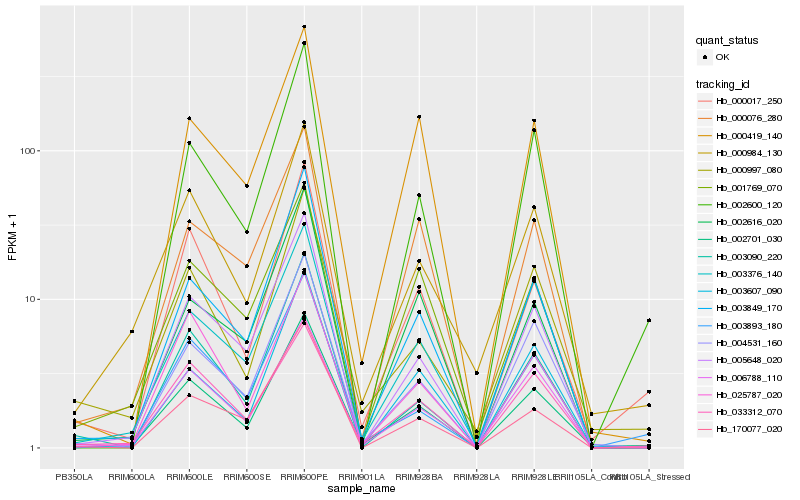
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002701_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 2 |
Hb_000017_250 |
0.0863431445 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 3 |
Hb_005648_020 |
0.0911918351 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM1 [Jatropha curcas] |
| 4 |
Hb_003849_170 |
0.0933744242 |
- |
- |
hypothetical protein JCGZ_17850 [Jatropha curcas] |
| 5 |
Hb_002600_120 |
0.0973967507 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 6 |
Hb_004531_160 |
0.098129279 |
- |
- |
PREDICTED: transcription factor bHLH68-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000997_080 |
0.1056863766 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000984_130 |
0.1058289311 |
- |
- |
tubulin [Ornithogalum longebracteatum] |
| 9 |
Hb_003893_180 |
0.1098872778 |
- |
- |
PREDICTED: uncharacterized protein LOC105648771 [Jatropha curcas] |
| 10 |
Hb_003607_090 |
0.1103144436 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 11 |
Hb_006788_110 |
0.1106063218 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 12 |
Hb_000419_140 |
0.1120453519 |
- |
- |
Aquaporin PIP2.1, putative [Ricinus communis] |
| 13 |
Hb_002616_020 |
0.1187723473 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 14 |
Hb_003090_220 |
0.1190232053 |
- |
- |
PREDICTED: copper transporter 4 [Jatropha curcas] |
| 15 |
Hb_003376_140 |
0.1196725204 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 16 |
Hb_025787_020 |
0.1204219264 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 17 |
Hb_170077_020 |
0.1206121519 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 18 |
Hb_000076_280 |
0.1207850482 |
- |
- |
PREDICTED: pectinesterase-like [Populus euphratica] |
| 19 |
Hb_033312_070 |
0.1220723318 |
- |
- |
hypothetical protein POPTR_0017s02960g [Populus trichocarpa] |
| 20 |
Hb_001769_070 |
0.1223545122 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like isoform X1 [Jatropha curcas] |