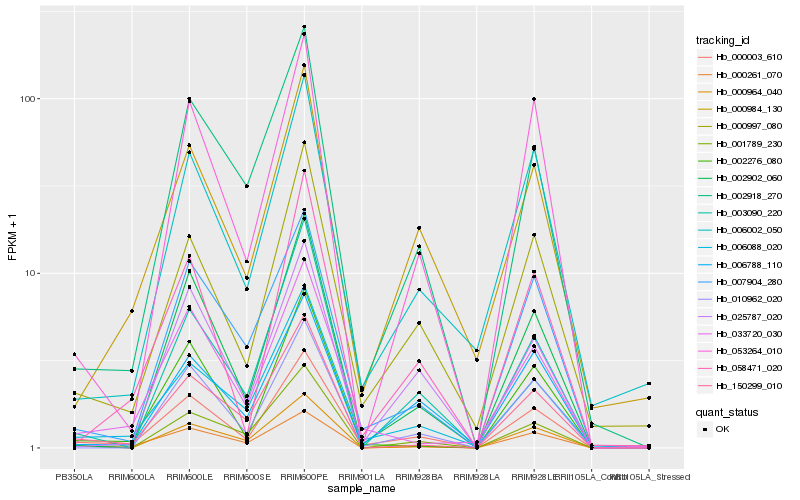
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_610 |
0.0 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase [Jatropha curcas] |
| 2 |
Hb_000997_080 |
0.0985430962 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_006088_020 |
0.0995719713 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 4 |
Hb_002918_270 |
0.1196930975 |
- |
- |
PREDICTED: protein E6-like [Populus euphratica] |
| 5 |
Hb_000984_130 |
0.1244268738 |
- |
- |
tubulin [Ornithogalum longebracteatum] |
| 6 |
Hb_000261_070 |
0.1247099636 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g06240-like [Jatropha curcas] |
| 7 |
Hb_002902_060 |
0.1267719003 |
- |
- |
PREDICTED: uncharacterized protein LOC105639824 [Jatropha curcas] |
| 8 |
Hb_053264_010 |
0.1271054645 |
- |
- |
beta glucosidase [Hevea brasiliensis] |
| 9 |
Hb_007904_280 |
0.1271208397 |
- |
- |
PREDICTED: uncharacterized protein LOC105644987 [Jatropha curcas] |
| 10 |
Hb_006788_110 |
0.128652444 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 11 |
Hb_058471_020 |
0.1292813277 |
- |
- |
PREDICTED: expansin-B3-like [Jatropha curcas] |
| 12 |
Hb_003090_220 |
0.1297442826 |
- |
- |
PREDICTED: copper transporter 4 [Jatropha curcas] |
| 13 |
Hb_010962_020 |
0.1308838955 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_0815310 [Ricinus communis] |
| 14 |
Hb_033720_030 |
0.1324394028 |
- |
- |
KDEL motif-containing protein 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_150299_010 |
0.1327569751 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 1-like [Tarenaya hassleriana] |
| 16 |
Hb_006002_050 |
0.1332996876 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 17 |
Hb_025787_020 |
0.1336855358 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 18 |
Hb_001789_230 |
0.1343259478 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000964_040 |
0.1354253149 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER isoform X2 [Jatropha curcas] |
| 20 |
Hb_002276_080 |
0.1357664798 |
- |
- |
Protein bem46, putative [Ricinus communis] |