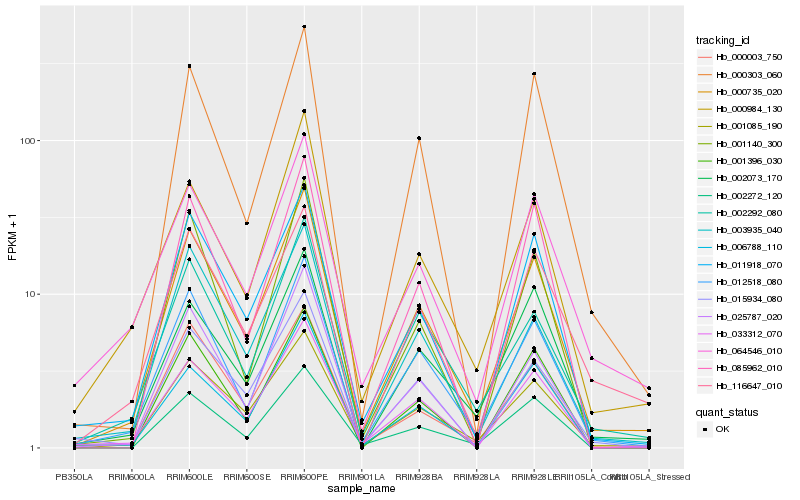
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000735_020 |
0.0 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 2 |
Hb_001085_190 |
0.053583102 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 3 |
Hb_012518_080 |
0.0657569095 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like [Jatropha curcas] |
| 4 |
Hb_011918_070 |
0.0826181058 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 5 |
Hb_033312_070 |
0.083991002 |
- |
- |
hypothetical protein POPTR_0017s02960g [Populus trichocarpa] |
| 6 |
Hb_002292_080 |
0.0929160679 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 7 |
Hb_006788_110 |
0.0948547775 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 8 |
Hb_025787_020 |
0.0970975521 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 9 |
Hb_000003_750 |
0.1002541613 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 10 |
Hb_015934_080 |
0.1008696463 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 11 |
Hb_003935_040 |
0.1012139298 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP [Jatropha curcas] |
| 12 |
Hb_002272_120 |
0.1016028272 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 13 |
Hb_000303_060 |
0.1016545752 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000984_130 |
0.1023132113 |
- |
- |
tubulin [Ornithogalum longebracteatum] |
| 15 |
Hb_064546_010 |
0.1086609416 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 16 |
Hb_002073_170 |
0.1089788063 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g27190 [Jatropha curcas] |
| 17 |
Hb_001396_030 |
0.1145703084 |
- |
- |
hypothetical protein POPTR_0001s13520g [Populus trichocarpa] |
| 18 |
Hb_001140_300 |
0.1154557068 |
- |
- |
PREDICTED: tubulin beta-1 chain [Cicer arietinum] |
| 19 |
Hb_116647_010 |
0.1158949773 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_085962_010 |
0.1162311029 |
- |
- |
hypothetical protein POPTR_0017s11380g [Populus trichocarpa] |