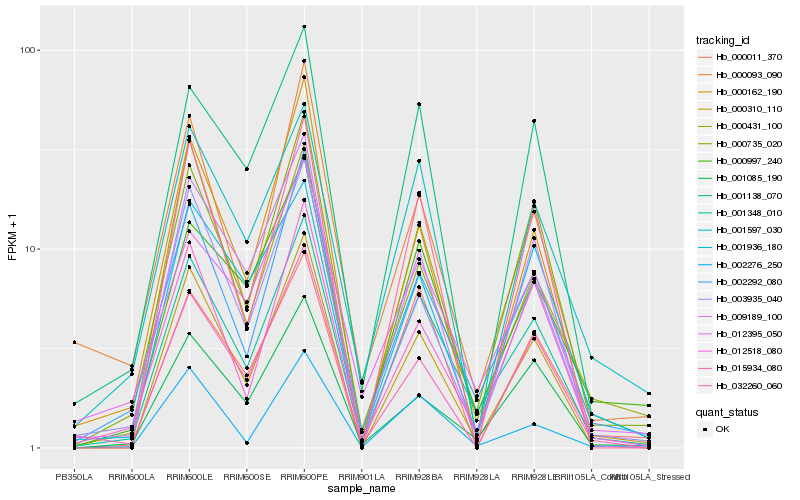
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001348_010 |
0.0 |
- |
- |
PREDICTED: lysM domain-containing GPI-anchored protein 1 [Jatropha curcas] |
| 2 |
Hb_002292_080 |
0.0861271186 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 3 |
Hb_000310_110 |
0.1035414068 |
- |
- |
PREDICTED: probable trehalose-phosphate phosphatase F isoform X1 [Jatropha curcas] |
| 4 |
Hb_000997_240 |
0.1107557783 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA4, putative [Ricinus communis] |
| 5 |
Hb_012395_050 |
0.1174358651 |
- |
- |
PREDICTED: uncharacterized protein LOC105632180 [Jatropha curcas] |
| 6 |
Hb_001138_070 |
0.1178978726 |
- |
- |
PREDICTED: D-3-phosphoglycerate dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_001085_190 |
0.122537937 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 8 |
Hb_001597_030 |
0.1235802554 |
- |
- |
PREDICTED: uncharacterized protein LOC104445128 isoform X1 [Eucalyptus grandis] |
| 9 |
Hb_012518_080 |
0.1240306193 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like [Jatropha curcas] |
| 10 |
Hb_009189_100 |
0.1240366716 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 11 |
Hb_003935_040 |
0.1258606311 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP [Jatropha curcas] |
| 12 |
Hb_000735_020 |
0.1316224802 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 13 |
Hb_002276_250 |
0.1330760473 |
- |
- |
PREDICTED: WD repeat and HMG-box DNA-binding protein 1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000162_190 |
0.1346740596 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 15 |
Hb_032260_060 |
0.1350119039 |
transcription factor |
TF Family: MYB-related |
Homeodomain-like superfamily protein [Theobroma cacao] |
| 16 |
Hb_015934_080 |
0.1377120011 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 17 |
Hb_001936_180 |
0.1379524892 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 18 |
Hb_000431_100 |
0.1382895117 |
- |
- |
ribonucleoside-diphosphate reductase small chain, putative [Ricinus communis] |
| 19 |
Hb_000011_370 |
0.1398838208 |
- |
- |
PREDICTED: uncharacterized protein LOC105631333 [Jatropha curcas] |
| 20 |
Hb_000093_090 |
0.1405612705 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |