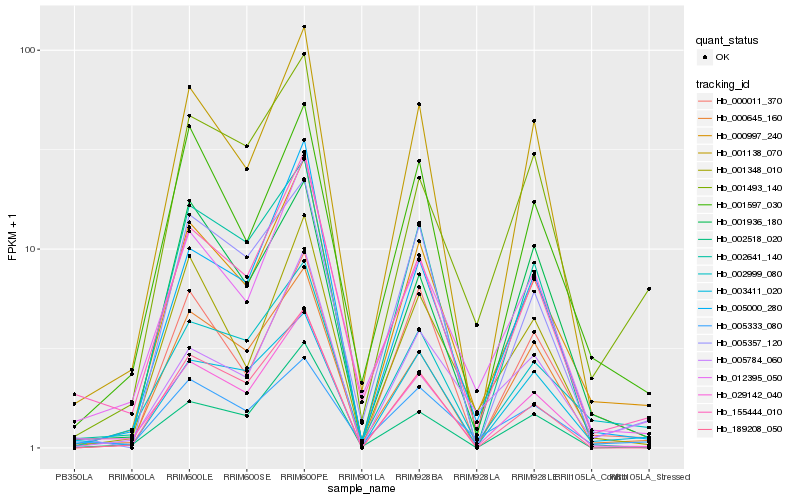
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000997_240 |
0.0 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA4, putative [Ricinus communis] |
| 2 |
Hb_002999_080 |
0.0915254538 |
transcription factor |
TF Family: G2-like |
PREDICTED: accessory gland protein Acp36DE [Jatropha curcas] |
| 3 |
Hb_001348_010 |
0.1107557783 |
- |
- |
PREDICTED: lysM domain-containing GPI-anchored protein 1 [Jatropha curcas] |
| 4 |
Hb_005000_280 |
0.1145937013 |
- |
- |
PREDICTED: uncharacterized protein LOC105637905 [Jatropha curcas] |
| 5 |
Hb_029142_040 |
0.1149498695 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 6 |
Hb_155444_010 |
0.115557772 |
- |
- |
- |
| 7 |
Hb_012395_050 |
0.1168467024 |
- |
- |
PREDICTED: uncharacterized protein LOC105632180 [Jatropha curcas] |
| 8 |
Hb_001138_070 |
0.1178894286 |
- |
- |
PREDICTED: D-3-phosphoglycerate dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_000645_160 |
0.1192520078 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 10 |
Hb_001597_030 |
0.119368518 |
- |
- |
PREDICTED: uncharacterized protein LOC104445128 isoform X1 [Eucalyptus grandis] |
| 11 |
Hb_001493_140 |
0.1220579648 |
- |
- |
hypothetical protein PHAVU_010G123100g [Phaseolus vulgaris] |
| 12 |
Hb_005784_060 |
0.1252041126 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 13 |
Hb_003411_020 |
0.1283135501 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 14 |
Hb_000011_370 |
0.1297755834 |
- |
- |
PREDICTED: uncharacterized protein LOC105631333 [Jatropha curcas] |
| 15 |
Hb_005333_080 |
0.134786042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_189208_050 |
0.1363780287 |
- |
- |
PREDICTED: uncharacterized protein LOC105642694 isoform X2 [Jatropha curcas] |
| 17 |
Hb_005357_120 |
0.1391730983 |
transcription factor |
TF Family: MYB-related |
- |
| 18 |
Hb_002641_140 |
0.1392025665 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 19 |
Hb_002518_020 |
0.1396379864 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 20 |
Hb_001936_180 |
0.1414568896 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |