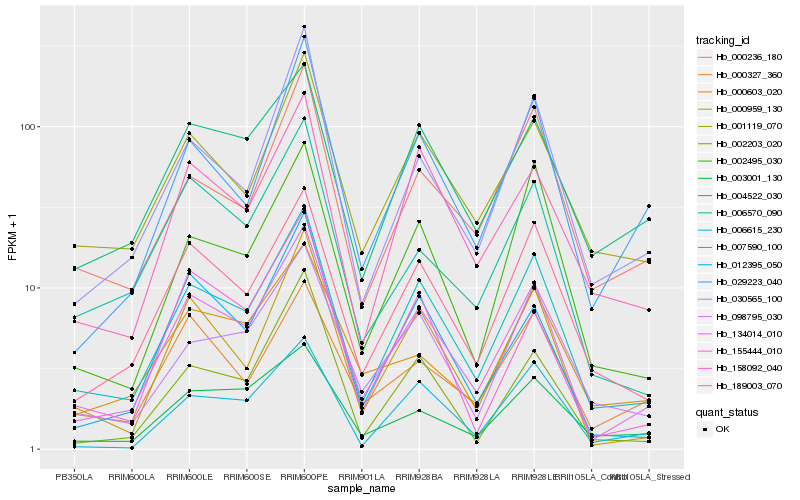
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_134014_010 |
0.0 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 2 |
Hb_006615_230 |
0.0706301438 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 3 |
Hb_189003_070 |
0.100842363 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 4 |
Hb_000959_130 |
0.1061611484 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 5 |
Hb_012395_050 |
0.1126741904 |
- |
- |
PREDICTED: uncharacterized protein LOC105632180 [Jatropha curcas] |
| 6 |
Hb_029223_040 |
0.1234423437 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 7 |
Hb_158092_040 |
0.1244271633 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 8 |
Hb_001119_070 |
0.1246874094 |
- |
- |
methylenetetrahydrofolate reductase, putative [Ricinus communis] |
| 9 |
Hb_006570_090 |
0.1252258439 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 10 |
Hb_030565_100 |
0.1289034953 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 11 |
Hb_004522_030 |
0.131370716 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 3 [Jatropha curcas] |
| 12 |
Hb_003001_130 |
0.1327223026 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_098795_030 |
0.1336706659 |
- |
- |
PREDICTED: metal tolerance protein 1-like [Vitis vinifera] |
| 14 |
Hb_007590_100 |
0.1350738575 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 15 |
Hb_000603_020 |
0.1356405057 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002203_020 |
0.1369634197 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_155444_010 |
0.1379170601 |
- |
- |
- |
| 18 |
Hb_002495_030 |
0.1411326443 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000327_360 |
0.1420818944 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_000236_180 |
0.1428073032 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |