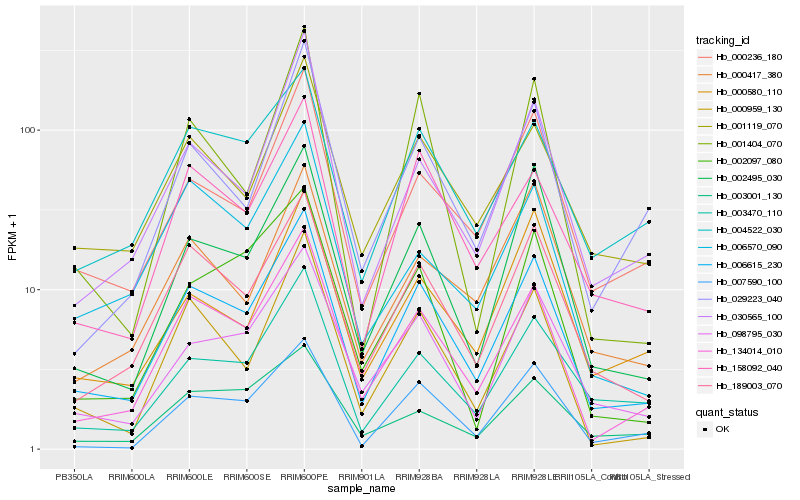
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006615_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 2 |
Hb_134014_010 |
0.0706301438 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 3 |
Hb_189003_070 |
0.0776442542 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 4 |
Hb_098795_030 |
0.0970083818 |
- |
- |
PREDICTED: metal tolerance protein 1-like [Vitis vinifera] |
| 5 |
Hb_001119_070 |
0.0995900167 |
- |
- |
methylenetetrahydrofolate reductase, putative [Ricinus communis] |
| 6 |
Hb_158092_040 |
0.1001450376 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 7 |
Hb_002495_030 |
0.1045649853 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000236_180 |
0.1072949039 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 9 |
Hb_003470_110 |
0.1107460023 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_000959_130 |
0.1122125703 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 11 |
Hb_007590_100 |
0.1137583477 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 12 |
Hb_004522_030 |
0.1161471509 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 3 [Jatropha curcas] |
| 13 |
Hb_030565_100 |
0.1227880329 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 14 |
Hb_029223_040 |
0.1271981908 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 15 |
Hb_003001_130 |
0.1307316615 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000417_380 |
0.1324660197 |
- |
- |
SecY protein transport family protein [Theobroma cacao] |
| 17 |
Hb_000580_110 |
0.1339796968 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3 [Jatropha curcas] |
| 18 |
Hb_006570_090 |
0.1364253885 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 19 |
Hb_001404_070 |
0.1373200232 |
- |
- |
sucrose synthase 4 [Hevea brasiliensis] |
| 20 |
Hb_002097_080 |
0.1385546966 |
- |
- |
phosphatidylinositol-4-phosphate 5-kinase, putative [Ricinus communis] |