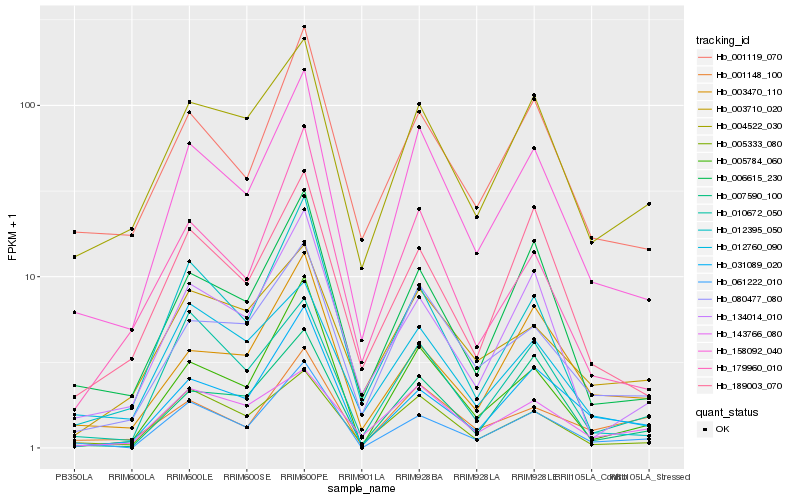
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_158092_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 2 |
Hb_080477_080 |
0.0986275069 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 isoform X1 [Gossypium raimondii] |
| 3 |
Hb_006615_230 |
0.1001450376 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 4 |
Hb_001119_070 |
0.1048199889 |
- |
- |
methylenetetrahydrofolate reductase, putative [Ricinus communis] |
| 5 |
Hb_005333_080 |
0.1094542643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004522_030 |
0.116156559 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 3 [Jatropha curcas] |
| 7 |
Hb_134014_010 |
0.1244271633 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 8 |
Hb_189003_070 |
0.1257436298 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 9 |
Hb_005784_060 |
0.1302525624 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 10 |
Hb_179960_010 |
0.1304216627 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Jatropha curcas] |
| 11 |
Hb_012760_090 |
0.1343751134 |
- |
- |
Aspartic proteinase-like protein 1 [Morus notabilis] |
| 12 |
Hb_061222_010 |
0.1354558398 |
- |
- |
hypothetical protein RCOM_0012650 [Ricinus communis] |
| 13 |
Hb_012395_050 |
0.1381030084 |
- |
- |
PREDICTED: uncharacterized protein LOC105632180 [Jatropha curcas] |
| 14 |
Hb_010672_050 |
0.1390186846 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X3 [Jatropha curcas] |
| 15 |
Hb_003470_110 |
0.1437682752 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_003710_020 |
0.1439164647 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007590_100 |
0.1447718522 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 18 |
Hb_143766_080 |
0.1469010985 |
- |
- |
PREDICTED: uncharacterized protein LOC105637797 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001148_100 |
0.1477131132 |
- |
- |
- |
| 20 |
Hb_031089_020 |
0.1478462269 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |