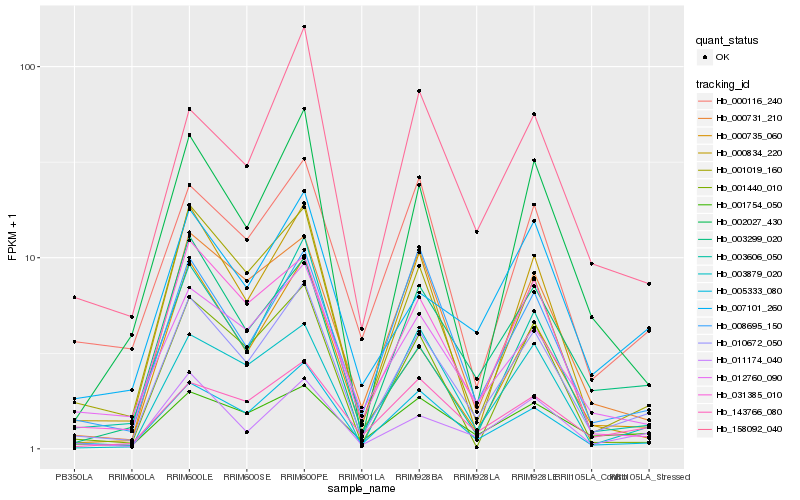
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010672_050 |
0.0 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X3 [Jatropha curcas] |
| 2 |
Hb_005333_080 |
0.0802642442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003299_020 |
0.1104558229 |
- |
- |
PREDICTED: uncharacterized protein LOC105649783 [Jatropha curcas] |
| 4 |
Hb_001019_160 |
0.1127738828 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_012760_090 |
0.1157047422 |
- |
- |
Aspartic proteinase-like protein 1 [Morus notabilis] |
| 6 |
Hb_000735_060 |
0.1188208362 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 7 |
Hb_011174_040 |
0.1208154542 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_031385_010 |
0.1208908046 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 9 |
Hb_000834_220 |
0.123091148 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 10 |
Hb_000731_210 |
0.1240375544 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_008695_150 |
0.1309272492 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 7 isoform X1 [Jatropha curcas] |
| 12 |
Hb_143766_080 |
0.130927316 |
- |
- |
PREDICTED: uncharacterized protein LOC105637797 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001754_050 |
0.1315489461 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 14 |
Hb_003606_050 |
0.1332906526 |
- |
- |
PREDICTED: beta-galactosidase 10 [Jatropha curcas] |
| 15 |
Hb_002027_430 |
0.1344500845 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 16 |
Hb_000116_240 |
0.135006208 |
transcription factor |
TF Family: HB |
homeobox-leucine zipper family protein [Populus trichocarpa] |
| 17 |
Hb_001440_010 |
0.1364951656 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 1 [Populus euphratica] |
| 18 |
Hb_003879_020 |
0.1376732658 |
- |
- |
PREDICTED: MATE efflux family protein 1-like [Jatropha curcas] |
| 19 |
Hb_158092_040 |
0.1390186846 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 20 |
Hb_007101_260 |
0.1400019164 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |