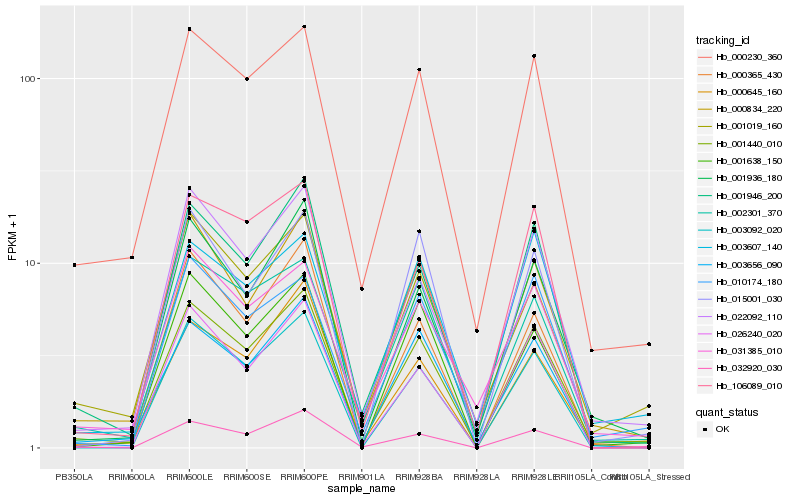
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001440_010 |
0.0 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 1 [Populus euphratica] |
| 2 |
Hb_000834_220 |
0.0748607923 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 3 |
Hb_001946_200 |
0.0754926298 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_000230_360 |
0.0818222051 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 5 |
Hb_003656_090 |
0.083364659 |
- |
- |
PREDICTED: protein Brevis radix-like 4 [Jatropha curcas] |
| 6 |
Hb_001936_180 |
0.084289378 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 7 |
Hb_022092_110 |
0.0886048075 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |
| 8 |
Hb_002301_370 |
0.0895475008 |
- |
- |
hypothetical protein JCGZ_21454 [Jatropha curcas] |
| 9 |
Hb_000645_160 |
0.0979012484 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 10 |
Hb_032920_030 |
0.1009263165 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 11 |
Hb_001019_160 |
0.1051768916 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001638_150 |
0.1062616463 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 13 |
Hb_015001_030 |
0.1070150014 |
- |
- |
inosine-uridine preferring nucleoside hydrolase family protein [Populus trichocarpa] |
| 14 |
Hb_003092_020 |
0.1096818494 |
- |
- |
PREDICTED: heptahelical transmembrane protein 1 [Jatropha curcas] |
| 15 |
Hb_106089_010 |
0.1109340478 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 16 |
Hb_010174_180 |
0.1110860709 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_003607_140 |
0.1113149411 |
- |
- |
3-ketoacyl-CoA thiolase B, putative [Ricinus communis] |
| 18 |
Hb_031385_010 |
0.1119463472 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 19 |
Hb_026240_020 |
0.1122529039 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 20 |
Hb_000365_430 |
0.1126875518 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g08570-like [Jatropha curcas] |