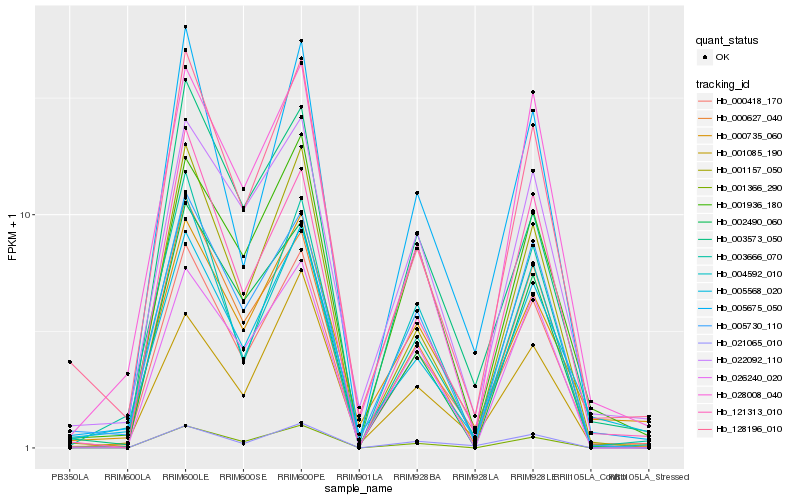
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000418_170 |
0.0 |
- |
- |
PREDICTED: ABC transporter C family member 12-like isoform X4 [Populus euphratica] |
| 2 |
Hb_000627_040 |
0.0889267492 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 3 |
Hb_022092_110 |
0.0895116241 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |
| 4 |
Hb_005730_110 |
0.0909906642 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 5 |
Hb_001936_180 |
0.0920349361 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 6 |
Hb_005675_050 |
0.0956657849 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 7 |
Hb_021065_010 |
0.0982812592 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like isoform X2 [Gossypium raimondii] |
| 8 |
Hb_128196_010 |
0.1017424214 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004592_010 |
0.1031750886 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b-like [Jatropha curcas] |
| 10 |
Hb_001366_290 |
0.1042371044 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_001157_050 |
0.1043074485 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 12 |
Hb_000735_060 |
0.1052696307 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 13 |
Hb_121313_010 |
0.106552162 |
- |
- |
PREDICTED: endoglucanase 24-like [Jatropha curcas] |
| 14 |
Hb_005568_020 |
0.1065887278 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 15 |
Hb_026240_020 |
0.1078918374 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 16 |
Hb_003666_070 |
0.1108157757 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |
| 17 |
Hb_003573_050 |
0.1115462332 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 18 |
Hb_002490_060 |
0.1123992822 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 19 |
Hb_001085_190 |
0.1130315923 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 20 |
Hb_028008_040 |
0.1131230144 |
- |
- |
conserved hypothetical protein [Ricinus communis] |