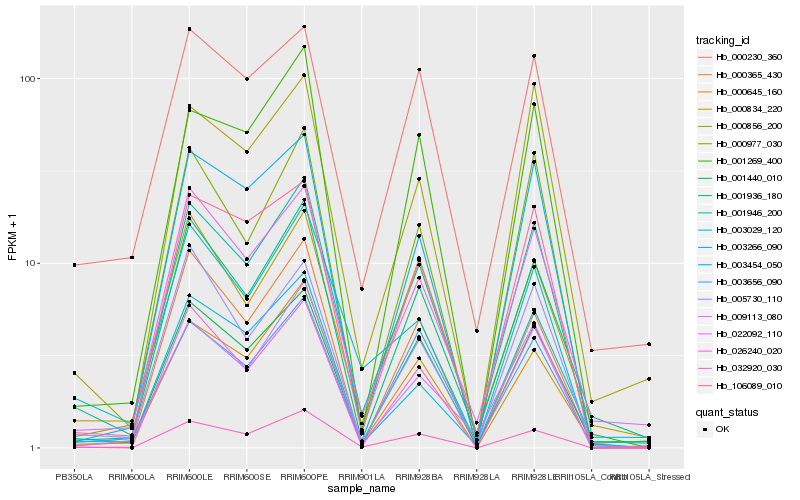
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001946_200 |
0.0 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 2 |
Hb_001440_010 |
0.0754926298 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 1 [Populus euphratica] |
| 3 |
Hb_022092_110 |
0.0811555307 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |
| 4 |
Hb_001936_180 |
0.0863895223 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 5 |
Hb_009113_080 |
0.0941957557 |
- |
- |
PREDICTED: deoxynucleoside triphosphate triphosphohydrolase SAMHD1 homolog [Jatropha curcas] |
| 6 |
Hb_026240_020 |
0.0942334319 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 7 |
Hb_032920_030 |
0.0947219047 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 8 |
Hb_106089_010 |
0.0981107795 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 9 |
Hb_000645_160 |
0.1009690591 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 10 |
Hb_000977_030 |
0.1020530783 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_003454_050 |
0.1041918791 |
- |
- |
PREDICTED: probable methyltransferase PMT14 [Jatropha curcas] |
| 12 |
Hb_000834_220 |
0.1055366951 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 13 |
Hb_000856_200 |
0.1058929466 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 14 |
Hb_005730_110 |
0.1083452549 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 15 |
Hb_001269_400 |
0.1083960774 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 16 |
Hb_003656_090 |
0.1121388334 |
- |
- |
PREDICTED: protein Brevis radix-like 4 [Jatropha curcas] |
| 17 |
Hb_000230_360 |
0.1136305544 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 18 |
Hb_003266_090 |
0.1137649255 |
- |
- |
PREDICTED: basic 7S globulin 2-like [Jatropha curcas] |
| 19 |
Hb_003029_120 |
0.1140804674 |
- |
- |
PREDICTED: uncharacterized protein LOC105642638 [Jatropha curcas] |
| 20 |
Hb_000365_430 |
0.1141042574 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g08570-like [Jatropha curcas] |