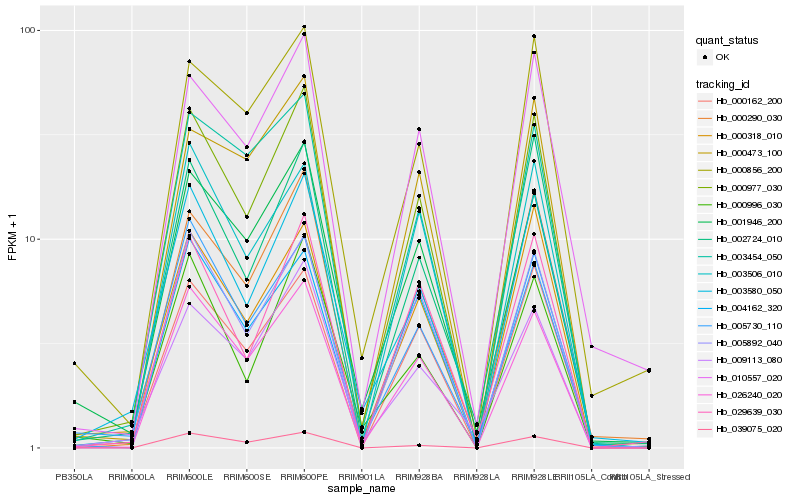
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000977_030 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_026240_020 |
0.0572995756 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 3 |
Hb_000290_030 |
0.0797773547 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 4 |
Hb_003580_050 |
0.081798784 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 5 |
Hb_000162_200 |
0.0828398259 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 6 |
Hb_005892_040 |
0.0880251507 |
- |
- |
hypothetical protein JCGZ_02368 [Jatropha curcas] |
| 7 |
Hb_002724_010 |
0.0920234343 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4-like [Citrus sinensis] |
| 8 |
Hb_029639_030 |
0.0921856126 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 2-like [Jatropha curcas] |
| 9 |
Hb_000318_010 |
0.0932082926 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 10 |
Hb_003506_010 |
0.1008161034 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 11 |
Hb_000996_030 |
0.1009976263 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g63430 isoform X1 [Jatropha curcas] |
| 12 |
Hb_009113_080 |
0.1013508908 |
- |
- |
PREDICTED: deoxynucleoside triphosphate triphosphohydrolase SAMHD1 homolog [Jatropha curcas] |
| 13 |
Hb_010557_020 |
0.1020138958 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 14 |
Hb_001946_200 |
0.1020530783 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 15 |
Hb_005730_110 |
0.1022418135 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 16 |
Hb_000473_100 |
0.1065752745 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 17 |
Hb_003454_050 |
0.1071984823 |
- |
- |
PREDICTED: probable methyltransferase PMT14 [Jatropha curcas] |
| 18 |
Hb_039075_020 |
0.1079997503 |
- |
- |
PREDICTED: uncharacterized protein LOC103331245 [Prunus mume] |
| 19 |
Hb_000856_200 |
0.1095656332 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 20 |
Hb_004162_320 |
0.1106731577 |
- |
- |
PREDICTED: uncharacterized protein LOC105628009 isoform X1 [Jatropha curcas] |