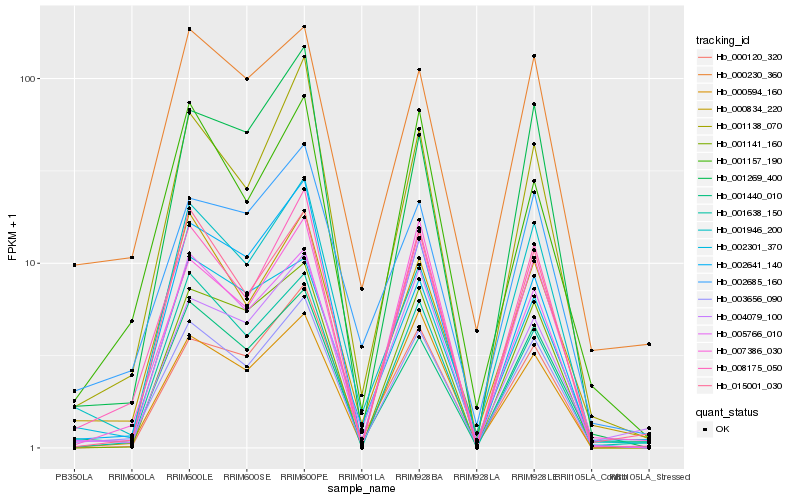
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003656_090 |
0.0 |
- |
- |
PREDICTED: protein Brevis radix-like 4 [Jatropha curcas] |
| 2 |
Hb_008175_050 |
0.0599371047 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 3 |
Hb_001440_010 |
0.083364659 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 1 [Populus euphratica] |
| 4 |
Hb_000834_220 |
0.0922765673 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 5 |
Hb_001138_070 |
0.093308835 |
- |
- |
PREDICTED: D-3-phosphoglycerate dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_015001_030 |
0.1034530458 |
- |
- |
inosine-uridine preferring nucleoside hydrolase family protein [Populus trichocarpa] |
| 7 |
Hb_001141_160 |
0.1056767397 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 8 |
Hb_000120_320 |
0.1079482765 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_000594_160 |
0.1087195165 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 10 |
Hb_002301_370 |
0.1091009198 |
- |
- |
hypothetical protein JCGZ_21454 [Jatropha curcas] |
| 11 |
Hb_000230_360 |
0.1099734463 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 12 |
Hb_001638_150 |
0.1105138977 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 13 |
Hb_005766_010 |
0.1105430365 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 14 |
Hb_002641_140 |
0.110801377 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 15 |
Hb_001946_200 |
0.1121388334 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_002685_160 |
0.1140471354 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 17 |
Hb_001157_190 |
0.1170466706 |
- |
- |
PREDICTED: protein YLS3 [Jatropha curcas] |
| 18 |
Hb_007386_030 |
0.1174422992 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 19 |
Hb_001269_400 |
0.1178435012 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 20 |
Hb_004079_100 |
0.1205649937 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |