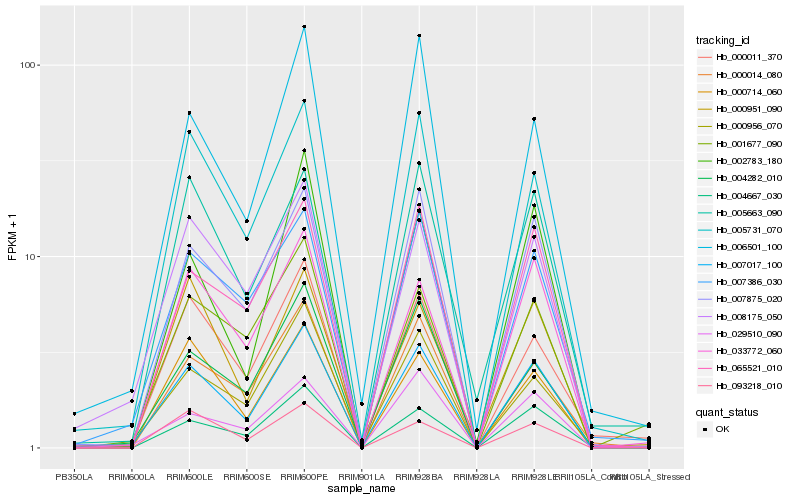
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007017_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649804 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007875_020 |
0.0820693423 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 3 |
Hb_005731_070 |
0.0935074701 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_008175_050 |
0.0988873999 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 5 |
Hb_065521_010 |
0.1037525675 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL5 [Jatropha curcas] |
| 6 |
Hb_007386_030 |
0.1049907908 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 7 |
Hb_000714_060 |
0.1144590122 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 1-like [Populus euphratica] |
| 8 |
Hb_033772_060 |
0.1145151899 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004667_030 |
0.1158898691 |
- |
- |
hypothetical protein VITISV_034568 [Vitis vinifera] |
| 10 |
Hb_000956_070 |
0.1168221238 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 11 |
Hb_000014_080 |
0.1173517439 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Jatropha curcas] |
| 12 |
Hb_006501_100 |
0.1186668868 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 13 |
Hb_004282_010 |
0.1191562172 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 14 |
Hb_029510_090 |
0.1252670241 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 15 |
Hb_005663_090 |
0.1256211974 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 7 [Jatropha curcas] |
| 16 |
Hb_002783_180 |
0.1266934469 |
- |
- |
Tubulin beta-6 chain [Gossypium arboreum] |
| 17 |
Hb_093218_010 |
0.1272571691 |
- |
- |
hypothetical protein POPTR_0001s05220g [Populus trichocarpa] |
| 18 |
Hb_000011_370 |
0.1290047347 |
- |
- |
PREDICTED: uncharacterized protein LOC105631333 [Jatropha curcas] |
| 19 |
Hb_000951_090 |
0.1292671435 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 20 |
Hb_001677_090 |
0.1334020496 |
- |
- |
PREDICTED: CASP-like protein 5B2 [Jatropha curcas] |