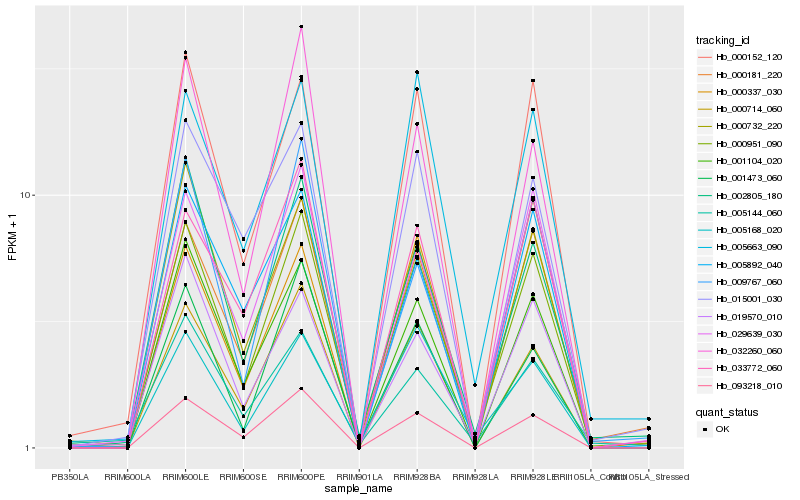
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000951_090 |
0.0 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 2 |
Hb_093218_010 |
0.0901529124 |
- |
- |
hypothetical protein POPTR_0001s05220g [Populus trichocarpa] |
| 3 |
Hb_001104_020 |
0.0903797228 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_000152_120 |
0.0927449016 |
- |
- |
alpha-l-fucosidase, putative [Ricinus communis] |
| 5 |
Hb_029639_030 |
0.0934901296 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 2-like [Jatropha curcas] |
| 6 |
Hb_000714_060 |
0.0940130134 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 1-like [Populus euphratica] |
| 7 |
Hb_002805_180 |
0.0993351088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000732_220 |
0.1010258603 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 9 |
Hb_000181_220 |
0.1094220931 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_005663_090 |
0.109636755 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 7 [Jatropha curcas] |
| 11 |
Hb_019570_010 |
0.1096546604 |
- |
- |
lyase, putative [Ricinus communis] |
| 12 |
Hb_001473_060 |
0.116885933 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 8 [Jatropha curcas] |
| 13 |
Hb_033772_060 |
0.117696697 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005144_060 |
0.1194727519 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 15 |
Hb_009767_060 |
0.1201800613 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 16 |
Hb_032260_060 |
0.1207138538 |
transcription factor |
TF Family: MYB-related |
Homeodomain-like superfamily protein [Theobroma cacao] |
| 17 |
Hb_005168_020 |
0.1207627055 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Jatropha curcas] |
| 18 |
Hb_000337_030 |
0.1215893571 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 19 |
Hb_015001_030 |
0.1233810329 |
- |
- |
inosine-uridine preferring nucleoside hydrolase family protein [Populus trichocarpa] |
| 20 |
Hb_005892_040 |
0.125162441 |
- |
- |
hypothetical protein JCGZ_02368 [Jatropha curcas] |