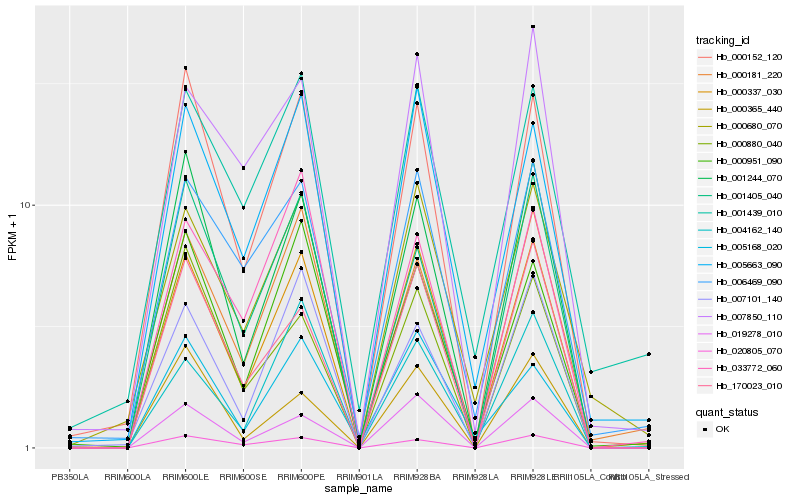
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000337_030 |
0.0 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 2 |
Hb_019278_010 |
0.087520422 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 3 |
Hb_000152_120 |
0.0954804361 |
- |
- |
alpha-l-fucosidase, putative [Ricinus communis] |
| 4 |
Hb_005663_090 |
0.0965240494 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 7 [Jatropha curcas] |
| 5 |
Hb_001244_070 |
0.099866232 |
- |
- |
hypothetical protein POPTR_0004s13470g [Populus trichocarpa] |
| 6 |
Hb_000181_220 |
0.1108504204 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_170023_010 |
0.1112494237 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 8 |
Hb_020805_070 |
0.1133419523 |
- |
- |
PREDICTED: MLO protein homolog 1-like [Jatropha curcas] |
| 9 |
Hb_006469_090 |
0.1152570988 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |
| 10 |
Hb_007850_110 |
0.1204263474 |
- |
- |
MADS-box transcription factor 3 [Hevea brasiliensis] |
| 11 |
Hb_000951_090 |
0.1215893571 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 12 |
Hb_000680_070 |
0.1231845241 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007101_140 |
0.1321728143 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |
| 14 |
Hb_004162_140 |
0.1355656179 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 15 |
Hb_005168_020 |
0.1357818836 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Jatropha curcas] |
| 16 |
Hb_001439_010 |
0.1376158311 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 5-like [Jatropha curcas] |
| 17 |
Hb_001405_040 |
0.1380067376 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Jatropha curcas] |
| 18 |
Hb_033772_060 |
0.1380443965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000880_040 |
0.1382482082 |
- |
- |
- |
| 20 |
Hb_000365_440 |
0.1390635602 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |