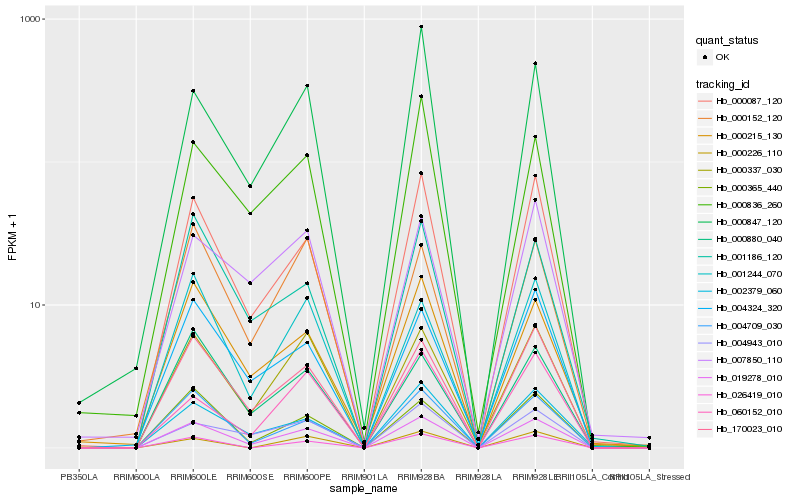
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019278_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 2 |
Hb_170023_010 |
0.0747873896 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 3 |
Hb_000087_120 |
0.0772868602 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 4 |
Hb_000337_030 |
0.087520422 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 5 |
Hb_004709_030 |
0.0907420987 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 6 |
Hb_000365_440 |
0.0989246877 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_002379_060 |
0.1034817284 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 8 |
Hb_001244_070 |
0.1154886732 |
- |
- |
hypothetical protein POPTR_0004s13470g [Populus trichocarpa] |
| 9 |
Hb_004324_320 |
0.1164875433 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 10 |
Hb_026419_010 |
0.116544871 |
- |
- |
PREDICTED: cyclin-D3-1-like [Jatropha curcas] |
| 11 |
Hb_000215_130 |
0.1176264289 |
- |
- |
PREDICTED: probable pectinesterase 53 [Jatropha curcas] |
| 12 |
Hb_000880_040 |
0.1235409038 |
- |
- |
- |
| 13 |
Hb_004943_010 |
0.1239371865 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 14 |
Hb_000836_260 |
0.128766445 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 15 |
Hb_000152_120 |
0.1289994919 |
- |
- |
alpha-l-fucosidase, putative [Ricinus communis] |
| 16 |
Hb_000847_120 |
0.1299797792 |
- |
- |
hypothetical protein JCGZ_15115 [Jatropha curcas] |
| 17 |
Hb_001186_120 |
0.1323145013 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 18 |
Hb_000226_110 |
0.1405055338 |
- |
- |
- |
| 19 |
Hb_007850_110 |
0.1422427736 |
- |
- |
MADS-box transcription factor 3 [Hevea brasiliensis] |
| 20 |
Hb_060152_010 |
0.142535612 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |