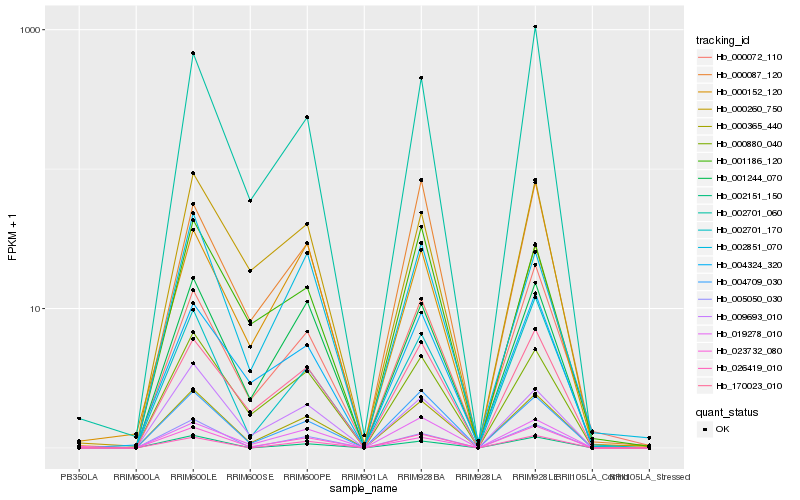
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000365_440 |
0.0 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_004709_030 |
0.0736732626 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 3 |
Hb_000880_040 |
0.079539945 |
- |
- |
- |
| 4 |
Hb_001244_070 |
0.0834484029 |
- |
- |
hypothetical protein POPTR_0004s13470g [Populus trichocarpa] |
| 5 |
Hb_170023_010 |
0.0839721019 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 6 |
Hb_005050_030 |
0.0896218866 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like [Jatropha curcas] |
| 7 |
Hb_019278_010 |
0.0989246877 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 8 |
Hb_009693_010 |
0.1034412257 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Gossypium raimondii] |
| 9 |
Hb_004324_320 |
0.1079798955 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 10 |
Hb_002701_170 |
0.1109832194 |
- |
- |
PREDICTED: cytochrome P450 85A [Jatropha curcas] |
| 11 |
Hb_002151_150 |
0.1114685036 |
- |
- |
PREDICTED: uncharacterized protein LOC102707106 [Oryza brachyantha] |
| 12 |
Hb_002851_070 |
0.1156108681 |
- |
- |
PREDICTED: uncharacterized protein LOC105646410 [Jatropha curcas] |
| 13 |
Hb_000260_750 |
0.1197167214 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 14 |
Hb_001186_120 |
0.1229663782 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 15 |
Hb_000072_110 |
0.1242078568 |
- |
- |
PREDICTED: U-box domain-containing protein 35 [Jatropha curcas] |
| 16 |
Hb_000087_120 |
0.1257499367 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 17 |
Hb_002701_060 |
0.1310847224 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000152_120 |
0.1315718214 |
- |
- |
alpha-l-fucosidase, putative [Ricinus communis] |
| 19 |
Hb_026419_010 |
0.1335531416 |
- |
- |
PREDICTED: cyclin-D3-1-like [Jatropha curcas] |
| 20 |
Hb_023732_080 |
0.134936543 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |