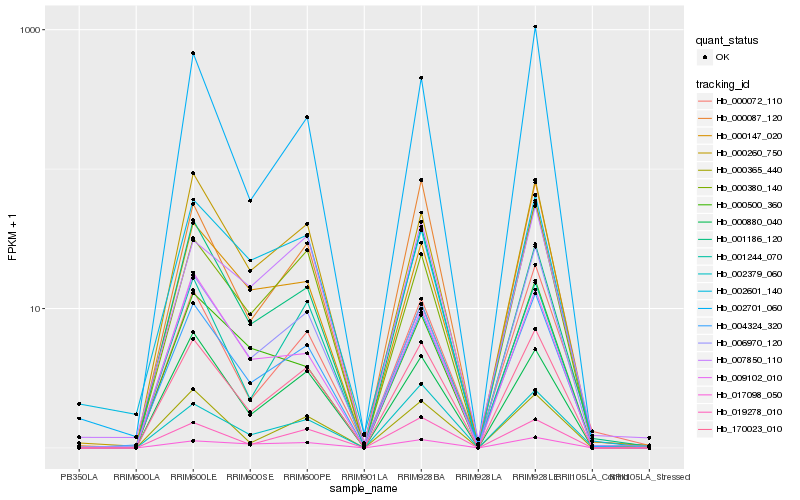
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004324_320 |
0.0 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 2 |
Hb_170023_010 |
0.0566559229 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 3 |
Hb_000147_020 |
0.0879580551 |
- |
- |
PREDICTED: uncharacterized protein LOC105638303 [Jatropha curcas] |
| 4 |
Hb_000260_750 |
0.0879724985 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 5 |
Hb_000072_110 |
0.0999676654 |
- |
- |
PREDICTED: U-box domain-containing protein 35 [Jatropha curcas] |
| 6 |
Hb_000880_040 |
0.1036132006 |
- |
- |
- |
| 7 |
Hb_000365_440 |
0.1079798955 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_006970_120 |
0.1081921501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000087_120 |
0.1124357403 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 10 |
Hb_019278_010 |
0.1164875433 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 11 |
Hb_000380_140 |
0.1168337747 |
- |
- |
PREDICTED: uncharacterized protein LOC105628069 [Jatropha curcas] |
| 12 |
Hb_002701_060 |
0.1171126847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_017098_050 |
0.1183729491 |
- |
- |
hypothetical protein POPTR_0001s03540g [Populus trichocarpa] |
| 14 |
Hb_009102_010 |
0.1186637318 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_002601_140 |
0.1191096677 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 16 |
Hb_001244_070 |
0.1214586999 |
- |
- |
hypothetical protein POPTR_0004s13470g [Populus trichocarpa] |
| 17 |
Hb_001186_120 |
0.1219557777 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 18 |
Hb_000500_360 |
0.1221984084 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_007850_110 |
0.126409256 |
- |
- |
MADS-box transcription factor 3 [Hevea brasiliensis] |
| 20 |
Hb_002379_060 |
0.1281814988 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |