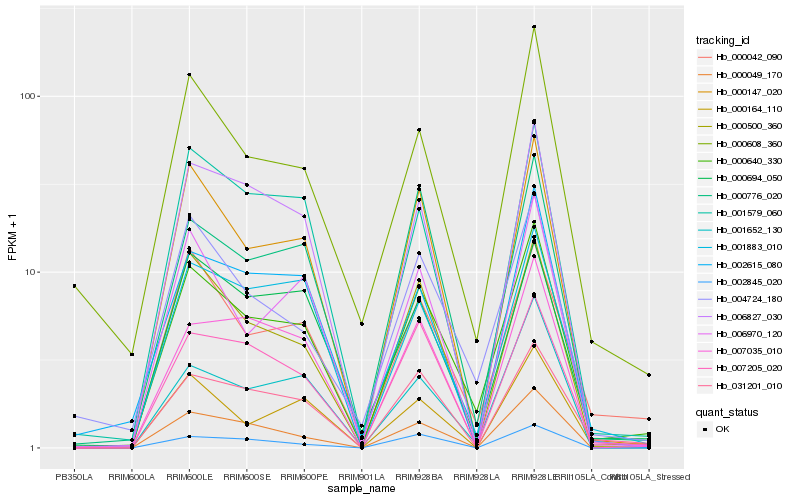
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000640_330 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000147_020 |
0.1073726845 |
- |
- |
PREDICTED: uncharacterized protein LOC105638303 [Jatropha curcas] |
| 3 |
Hb_004724_180 |
0.1150590531 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 1 [Jatropha curcas] |
| 4 |
Hb_000042_090 |
0.1190907827 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS2, putative [Ricinus communis] |
| 5 |
Hb_000776_020 |
0.1275669805 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_25594 [Jatropha curcas] |
| 6 |
Hb_000500_360 |
0.1281550314 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_000608_360 |
0.1282942602 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 8 |
Hb_006827_030 |
0.1290151813 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 9 |
Hb_031201_010 |
0.1299455998 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_000049_170 |
0.1310651722 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001579_060 |
0.1315279881 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 12 |
Hb_001652_130 |
0.1340674295 |
- |
- |
hypothetical protein B456_002G193900 [Gossypium raimondii] |
| 13 |
Hb_006970_120 |
0.13585538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001883_010 |
0.1369727209 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 15 |
Hb_007205_020 |
0.1372488619 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 16 |
Hb_000164_110 |
0.1397869873 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s44130g [Populus trichocarpa] |
| 17 |
Hb_000694_050 |
0.139850536 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 18 |
Hb_002615_080 |
0.1451668928 |
- |
- |
PREDICTED: non-specific phospholipase C2 [Jatropha curcas] |
| 19 |
Hb_002845_020 |
0.1454836982 |
- |
- |
hypothetical protein POPTR_0012s14920g [Populus trichocarpa] |
| 20 |
Hb_007035_010 |
0.1462106165 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |