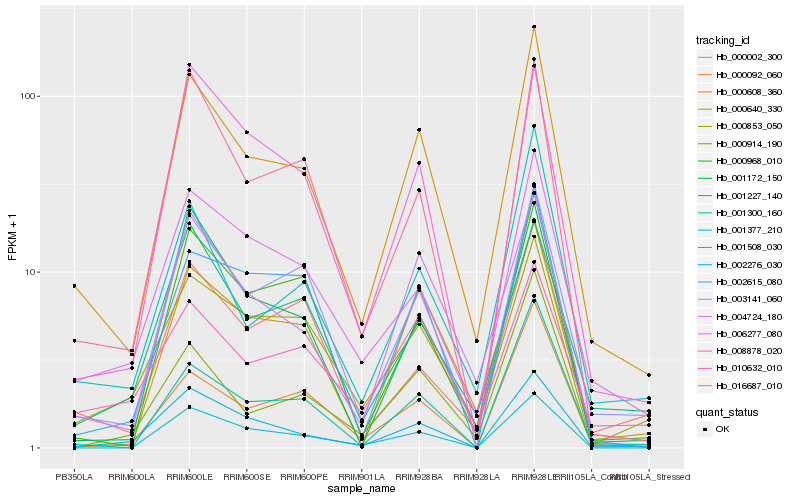
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000608_360 |
0.0 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 2 |
Hb_004724_180 |
0.110749894 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 1 [Jatropha curcas] |
| 3 |
Hb_003141_060 |
0.1109557749 |
transcription factor |
TF Family: TRAF |
PREDICTED: ETO1-like protein 1 [Jatropha curcas] |
| 4 |
Hb_000640_330 |
0.1282942602 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006277_080 |
0.1303600944 |
- |
- |
PREDICTED: uncharacterized protein LOC105649701 [Jatropha curcas] |
| 6 |
Hb_001300_160 |
0.1304047404 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like [Jatropha curcas] |
| 7 |
Hb_002276_030 |
0.1325146592 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 8 |
Hb_001377_210 |
0.1329029122 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 9 |
Hb_000968_010 |
0.1407617887 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000853_050 |
0.1412951321 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_016687_010 |
0.1444599268 |
- |
- |
PREDICTED: uncharacterized protein LOC105637401 [Jatropha curcas] |
| 12 |
Hb_000092_060 |
0.1461115789 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 13 |
Hb_001508_030 |
0.1478673046 |
- |
- |
hypothetical protein VITISV_029808 [Vitis vinifera] |
| 14 |
Hb_000914_190 |
0.152045541 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_002615_080 |
0.1522522818 |
- |
- |
PREDICTED: non-specific phospholipase C2 [Jatropha curcas] |
| 16 |
Hb_010632_010 |
0.15269677 |
- |
- |
PREDICTED: bifunctional dethiobiotin synthetase/7,8-diamino-pelargonic acid aminotransferase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_001172_150 |
0.1532535535 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 18 |
Hb_001227_140 |
0.1544898571 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 19 |
Hb_008878_020 |
0.1551581598 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member H1-like [Populus euphratica] |
| 20 |
Hb_000002_300 |
0.1551638536 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |