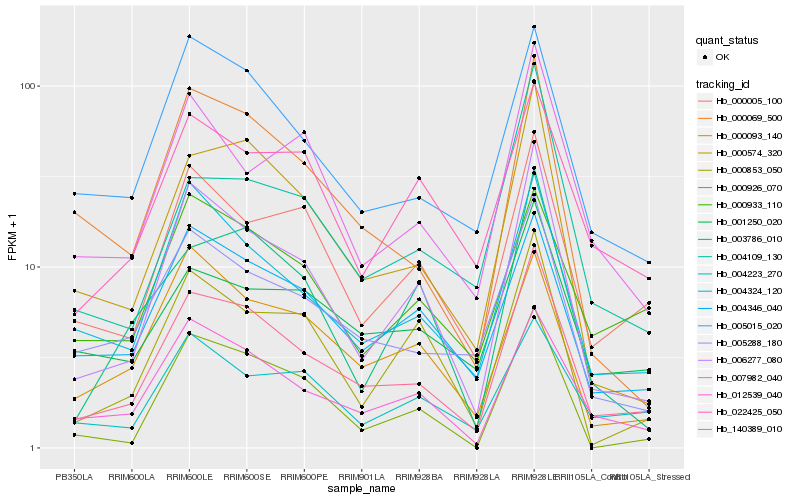
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_140389_010 |
0.0 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 2 |
Hb_006277_080 |
0.1048418494 |
- |
- |
PREDICTED: uncharacterized protein LOC105649701 [Jatropha curcas] |
| 3 |
Hb_000574_320 |
0.1134899566 |
- |
- |
hypothetical protein CICLE_v10024937mg [Citrus clementina] |
| 4 |
Hb_012539_040 |
0.1160844977 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_07765 [Jatropha curcas] |
| 5 |
Hb_005015_020 |
0.1240864578 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_004324_120 |
0.1243135538 |
- |
- |
ATP-dependent zinc metalloprotease YME1L1 [Gossypium arboreum] |
| 7 |
Hb_005288_180 |
0.1303121951 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04760, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_000005_100 |
0.1324734324 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_004346_040 |
0.1349636381 |
- |
- |
hypothetical protein POPTR_0016s06630g [Populus trichocarpa] |
| 10 |
Hb_000069_500 |
0.1397190764 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 4, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001250_020 |
0.1505113215 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_007982_040 |
0.1522402264 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 13 |
Hb_004109_130 |
0.1545318887 |
- |
- |
PREDICTED: translation initiation factor IF-2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000093_140 |
0.1553283399 |
- |
- |
PREDICTED: fructokinase-like 2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_004223_270 |
0.1564249568 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000926_070 |
0.158303062 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000933_110 |
0.1587663581 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 18 |
Hb_022425_050 |
0.1592436144 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 19 |
Hb_000853_050 |
0.1599558435 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_003786_010 |
0.1611057832 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |