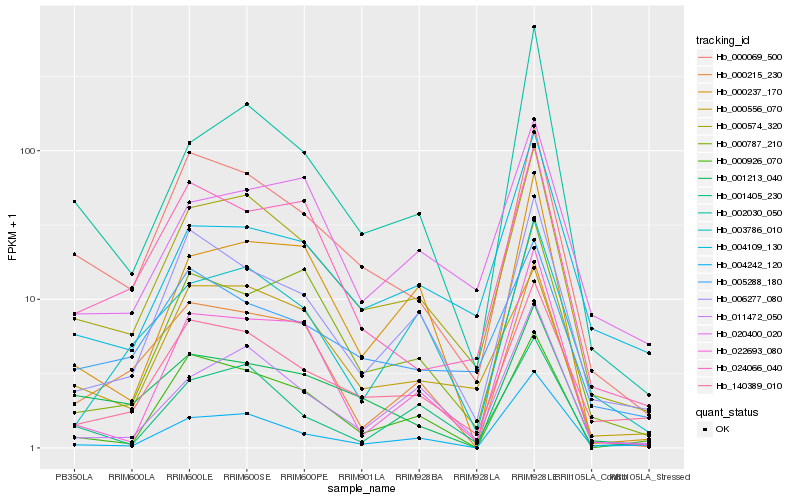
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000574_320 |
0.0 |
- |
- |
hypothetical protein CICLE_v10024937mg [Citrus clementina] |
| 2 |
Hb_140389_010 |
0.1134899566 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 3 |
Hb_000069_500 |
0.115262032 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 4, chloroplastic [Jatropha curcas] |
| 4 |
Hb_011472_050 |
0.1352823912 |
- |
- |
- |
| 5 |
Hb_006277_080 |
0.1362649639 |
- |
- |
PREDICTED: uncharacterized protein LOC105649701 [Jatropha curcas] |
| 6 |
Hb_000556_070 |
0.1398409783 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At2g45590 [Jatropha curcas] |
| 7 |
Hb_002030_050 |
0.1413857795 |
- |
- |
classical arabinogalactan protein 7 precursor [Jatropha curcas] |
| 8 |
Hb_001405_230 |
0.1460352026 |
- |
- |
STELAR K+ outward rectifier isoform 2 [Theobroma cacao] |
| 9 |
Hb_004242_120 |
0.1474439673 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 10 |
Hb_000215_230 |
0.1511211771 |
- |
- |
PREDICTED: uncharacterized protein LOC105637821 [Jatropha curcas] |
| 11 |
Hb_000787_210 |
0.1516937701 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000926_070 |
0.1520618036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004109_130 |
0.1528160801 |
- |
- |
PREDICTED: translation initiation factor IF-2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000237_170 |
0.1529552343 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 6 [Jatropha curcas] |
| 15 |
Hb_020400_020 |
0.1541557751 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 16 |
Hb_003786_010 |
0.1545974403 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_024066_040 |
0.156509894 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 18 |
Hb_005288_180 |
0.1571777516 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04760, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_001213_040 |
0.1583061195 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 20 |
Hb_022693_080 |
0.1617109473 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |