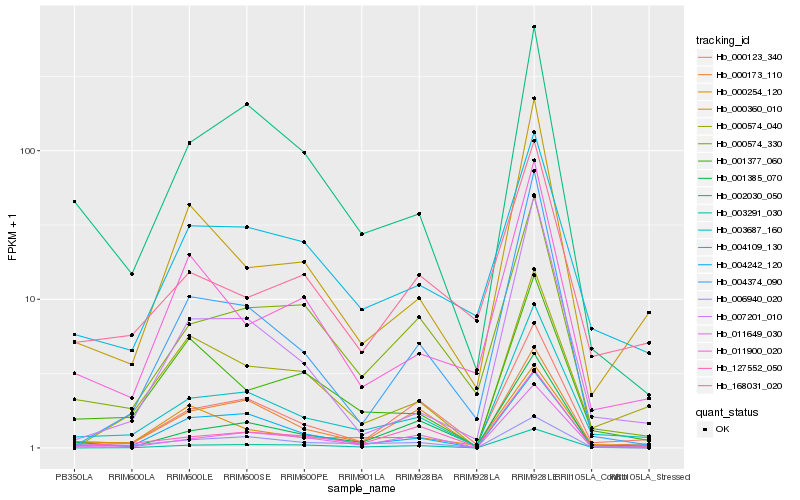
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003687_160 |
0.0 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 2 |
Hb_000123_340 |
0.1258535564 |
- |
- |
PREDICTED: uncharacterized protein LOC105789586 [Gossypium raimondii] |
| 3 |
Hb_004242_120 |
0.1341428558 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 4 |
Hb_000360_010 |
0.1453652358 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_004109_130 |
0.1462029333 |
- |
- |
PREDICTED: translation initiation factor IF-2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_011649_030 |
0.1469424939 |
- |
- |
hypothetical protein [Vitis hybrid cultivar] |
| 7 |
Hb_127552_050 |
0.1481543558 |
- |
- |
hypothetical protein VITISV_030752 [Vitis vinifera] |
| 8 |
Hb_001385_070 |
0.1486918444 |
- |
- |
hypothetical protein VITISV_019194 [Vitis vinifera] |
| 9 |
Hb_003291_030 |
0.1500282157 |
- |
- |
Gag-pol protein, putative [Solanum demissum] |
| 10 |
Hb_006940_020 |
0.1522882402 |
- |
- |
polyprotein [Oryza australiensis] |
| 11 |
Hb_000173_110 |
0.1537249947 |
- |
- |
putative RNA-directed DNA polymerase (Reverse transcriptase) [Malus domestica] |
| 12 |
Hb_000574_330 |
0.1546405341 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 13 |
Hb_002030_050 |
0.1550472161 |
- |
- |
classical arabinogalactan protein 7 precursor [Jatropha curcas] |
| 14 |
Hb_007201_010 |
0.1570841933 |
- |
- |
PREDICTED: uncharacterized protein LOC105631226 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000254_120 |
0.1607248259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_168031_020 |
0.1631724255 |
- |
- |
PREDICTED: uncharacterized protein LOC105646684 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000574_040 |
0.1688176604 |
- |
- |
PREDICTED: triphosphate tunel metalloenzyme 3-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_004374_090 |
0.1702461837 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 19 |
Hb_011900_020 |
0.1709678116 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16390, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001377_060 |
0.1712467914 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01030, mitochondrial [Jatropha curcas] |