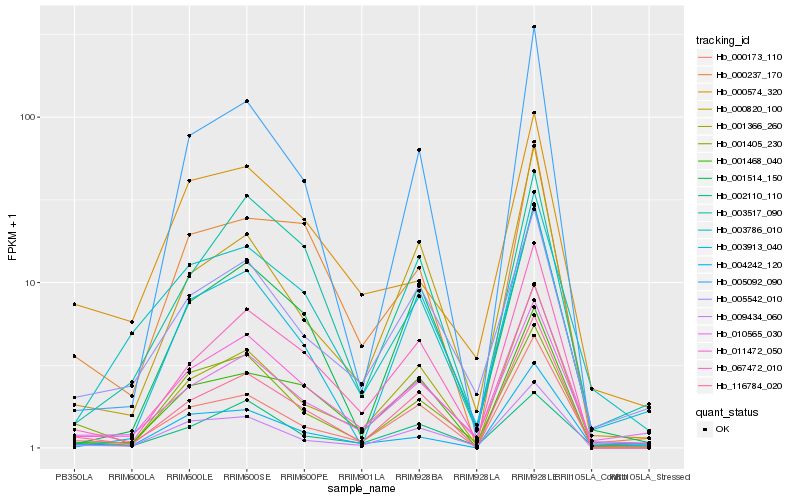
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011472_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_010565_030 |
0.0761710496 |
- |
- |
hypothetical protein VITISV_006810 [Vitis vinifera] |
| 3 |
Hb_005542_010 |
0.1063235418 |
- |
- |
Uncharacterized protein TCM_021447 [Theobroma cacao] |
| 4 |
Hb_067472_010 |
0.1090103076 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 5 |
Hb_001366_260 |
0.1101035963 |
- |
- |
T4.15 [Malus x robusta] |
| 6 |
Hb_000173_110 |
0.1139244448 |
- |
- |
putative RNA-directed DNA polymerase (Reverse transcriptase) [Malus domestica] |
| 7 |
Hb_009434_060 |
0.1173293161 |
- |
- |
T4.15 [Malus x robusta] |
| 8 |
Hb_116784_020 |
0.1189224739 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_001514_150 |
0.1192243993 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 10 |
Hb_001405_230 |
0.1260646799 |
- |
- |
STELAR K+ outward rectifier isoform 2 [Theobroma cacao] |
| 11 |
Hb_000820_100 |
0.1280732292 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |
| 12 |
Hb_003786_010 |
0.1311300575 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_003517_090 |
0.131427347 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_004242_120 |
0.131816024 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 15 |
Hb_002110_110 |
0.1319872247 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 16 |
Hb_000574_320 |
0.1352823912 |
- |
- |
hypothetical protein CICLE_v10024937mg [Citrus clementina] |
| 17 |
Hb_000237_170 |
0.1353827588 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 6 [Jatropha curcas] |
| 18 |
Hb_001468_040 |
0.1359821009 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 19 |
Hb_005092_090 |
0.1410968429 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 20 |
Hb_003913_040 |
0.1420330841 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |