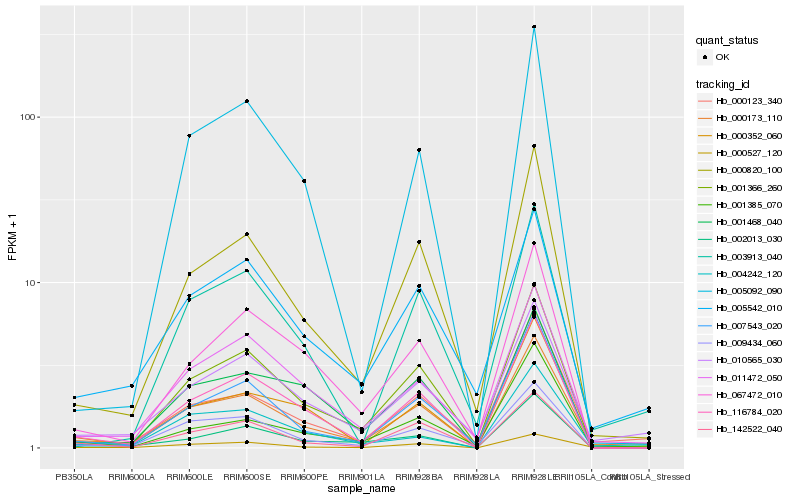
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000173_110 |
0.0 |
- |
- |
putative RNA-directed DNA polymerase (Reverse transcriptase) [Malus domestica] |
| 2 |
Hb_000123_340 |
0.0964639276 |
- |
- |
PREDICTED: uncharacterized protein LOC105789586 [Gossypium raimondii] |
| 3 |
Hb_001366_260 |
0.0983446924 |
- |
- |
T4.15 [Malus x robusta] |
| 4 |
Hb_000820_100 |
0.0990336043 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |
| 5 |
Hb_010565_030 |
0.1019773492 |
- |
- |
hypothetical protein VITISV_006810 [Vitis vinifera] |
| 6 |
Hb_116784_020 |
0.105032192 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_011472_050 |
0.1139244448 |
- |
- |
- |
| 8 |
Hb_000352_060 |
0.1180024989 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |
| 9 |
Hb_007543_020 |
0.1197246135 |
- |
- |
PREDICTED: uncharacterized protein LOC104229533, partial [Nicotiana sylvestris] |
| 10 |
Hb_000527_120 |
0.1227570633 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 11 |
Hb_009434_060 |
0.1275597302 |
- |
- |
T4.15 [Malus x robusta] |
| 12 |
Hb_004242_120 |
0.1310149302 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 13 |
Hb_142522_040 |
0.1332219153 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_005542_010 |
0.1364770234 |
- |
- |
Uncharacterized protein TCM_021447 [Theobroma cacao] |
| 15 |
Hb_002013_030 |
0.1375636141 |
- |
- |
PREDICTED: uncharacterized protein LOC104799692 [Tarenaya hassleriana] |
| 16 |
Hb_003913_040 |
0.1384665279 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 17 |
Hb_001468_040 |
0.1413020867 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 18 |
Hb_067472_010 |
0.1422623305 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 19 |
Hb_001385_070 |
0.1431999317 |
- |
- |
hypothetical protein VITISV_019194 [Vitis vinifera] |
| 20 |
Hb_005092_090 |
0.1447970201 |
- |
- |
cold regulated protein [Manihot esculenta] |