| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
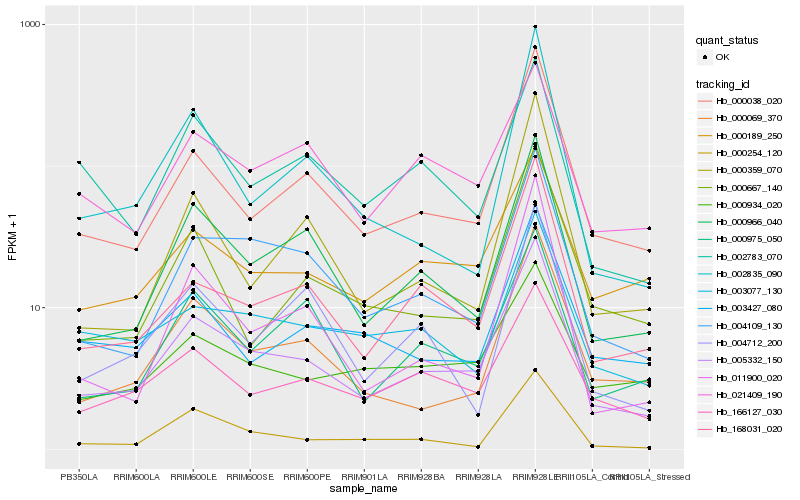
Hb_005332_150 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46610 [Jatropha curcas] |
| 2 |
Hb_000038_020 |
0.0990985708 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_004109_130 |
0.1087413492 |
- |
- |
PREDICTED: translation initiation factor IF-2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000966_040 |
0.1096442664 |
- |
- |
hypothetical protein JCGZ_01642 [Jatropha curcas] |
| 5 |
Hb_168031_020 |
0.119729262 |
- |
- |
PREDICTED: uncharacterized protein LOC105646684 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000069_370 |
0.1225947388 |
- |
- |
- |
| 7 |
Hb_166127_030 |
0.1301327592 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000189_250 |
0.1349004666 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000975_050 |
0.1413089488 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000254_120 |
0.1430967001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000667_140 |
0.1449995258 |
- |
- |
PREDICTED: uridine kinase-like protein 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003077_130 |
0.1450477087 |
- |
- |
hypothetical protein JCGZ_22476 [Jatropha curcas] |
| 13 |
Hb_003427_080 |
0.145561139 |
- |
- |
PREDICTED: probable alanine--tRNA ligase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_011900_020 |
0.1482413949 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16390, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002835_090 |
0.1503559949 |
- |
- |
RecName: Full=Elongation factor Tu, chloroplastic; Short=EF-Tu; Flags: Precursor [Glycine max] |
| 16 |
Hb_021409_190 |
0.1527252337 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 17 |
Hb_000934_020 |
0.1527842628 |
- |
- |
PREDICTED: uncharacterized protein LOC105629153 [Jatropha curcas] |
| 18 |
Hb_000359_070 |
0.1556386337 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002783_070 |
0.1582702339 |
- |
- |
PREDICTED: endoplasmin homolog [Jatropha curcas] |
| 20 |
Hb_004712_200 |
0.1583314923 |
- |
- |
hypothetical protein CICLE_v10010292mg [Citrus clementina] |