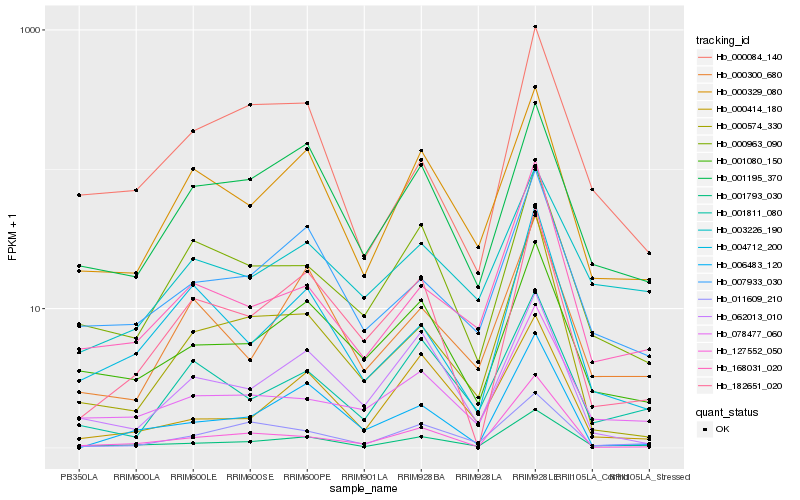
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001793_030 |
0.0 |
- |
- |
Cysteine-rich RLK (RECEPTOR-like protein kinase) 8 [Theobroma cacao] |
| 2 |
Hb_007933_030 |
0.1275699429 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 3 |
Hb_168031_020 |
0.1319363738 |
- |
- |
PREDICTED: uncharacterized protein LOC105646684 isoform X2 [Jatropha curcas] |
| 4 |
Hb_078477_060 |
0.133020474 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 5 |
Hb_001080_150 |
0.1338671076 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 6 |
Hb_000329_080 |
0.1353497446 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000414_180 |
0.153026317 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Vitis vinifera] |
| 8 |
Hb_000574_330 |
0.1631170212 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 9 |
Hb_001195_370 |
0.1643154039 |
- |
- |
- |
| 10 |
Hb_000300_680 |
0.1644272406 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000963_090 |
0.1649808436 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 12 |
Hb_011609_210 |
0.1660215576 |
- |
- |
Uncharacterized protein TCM_011393 [Theobroma cacao] |
| 13 |
Hb_001811_080 |
0.1660664413 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 14 |
Hb_000084_140 |
0.1676036126 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 15 |
Hb_003226_190 |
0.1684767356 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 16 |
Hb_004712_200 |
0.1697821374 |
- |
- |
hypothetical protein CICLE_v10010292mg [Citrus clementina] |
| 17 |
Hb_127552_050 |
0.1721628109 |
- |
- |
hypothetical protein VITISV_030752 [Vitis vinifera] |
| 18 |
Hb_006483_120 |
0.1743094845 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_182651_020 |
0.1760519366 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_062013_010 |
0.1764105232 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |