| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
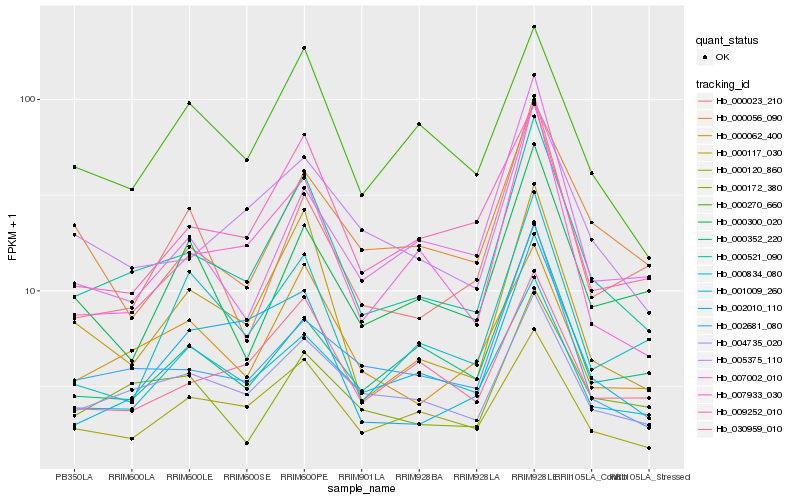
Hb_000521_090 |
0.0 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000117_030 |
0.1137114338 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 3 |
Hb_002681_080 |
0.1178221365 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004735_020 |
0.1197799856 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 5 |
Hb_007933_030 |
0.1202658936 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 6 |
Hb_000062_400 |
0.1267857546 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005375_110 |
0.1292428242 |
- |
- |
metal tolerance protein 1 [Populus trichocarpa x Populus deltoides] |
| 8 |
Hb_000120_860 |
0.1313607344 |
- |
- |
nucellin, putative [Ricinus communis] |
| 9 |
Hb_009252_010 |
0.1339101984 |
- |
- |
PREDICTED: kinesin light chain [Jatropha curcas] |
| 10 |
Hb_000172_380 |
0.1345442811 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 11 |
Hb_030959_010 |
0.1368646699 |
- |
- |
cullin, putative [Ricinus communis] |
| 12 |
Hb_000352_220 |
0.1390345184 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000270_660 |
0.1413097032 |
- |
- |
purine permease, putative [Ricinus communis] |
| 14 |
Hb_002010_110 |
0.1465666164 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 15 |
Hb_001009_260 |
0.1502774165 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_007002_010 |
0.1509305219 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 17 |
Hb_000056_090 |
0.1519892317 |
- |
- |
UDP-galactose transporter 3 isoform 1 [Theobroma cacao] |
| 18 |
Hb_000834_080 |
0.1525281759 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 19 |
Hb_000300_020 |
0.1540204394 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000023_210 |
0.1555343077 |
- |
- |
PREDICTED: protein TIC 55, chloroplastic [Jatropha curcas] |