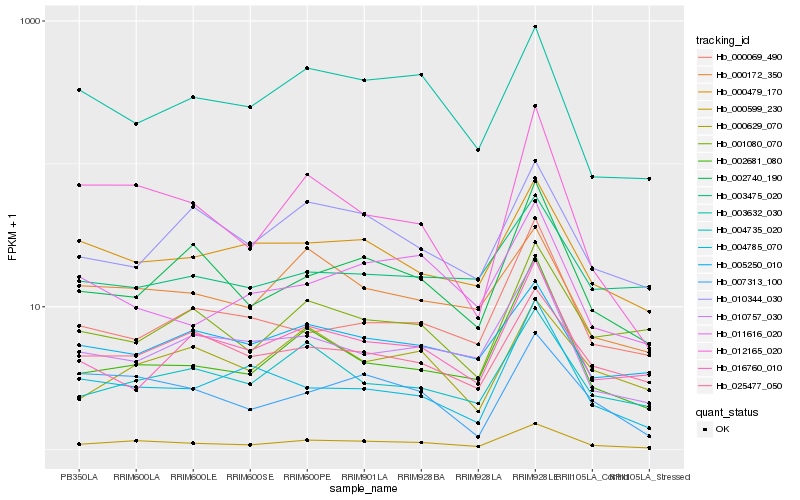
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000599_230 |
0.0 |
- |
- |
- |
| 2 |
Hb_004735_020 |
0.1174320139 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 3 |
Hb_002681_080 |
0.1235907124 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002740_190 |
0.124570877 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_010757_030 |
0.1264826042 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000479_170 |
0.1275336268 |
- |
- |
chaperone clpb, putative [Ricinus communis] |
| 7 |
Hb_025477_050 |
0.1323343984 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_000172_350 |
0.1337700341 |
- |
- |
PREDICTED: uncharacterized protein LOC105650963 [Jatropha curcas] |
| 9 |
Hb_007313_100 |
0.1386694472 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS2, putative [Ricinus communis] |
| 10 |
Hb_012165_020 |
0.1400506695 |
- |
- |
hypothetical protein POPTR_0007s05770g [Populus trichocarpa] |
| 11 |
Hb_005250_010 |
0.1413547985 |
- |
- |
hypothetical protein JCGZ_15339 [Jatropha curcas] |
| 12 |
Hb_003632_030 |
0.1434865093 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 13 |
Hb_001080_070 |
0.1445798859 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 14 |
Hb_010344_030 |
0.1460207956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_011616_020 |
0.1467274469 |
- |
- |
hypothetical protein CICLE_v10004968mg [Citrus clementina] |
| 16 |
Hb_000629_070 |
0.1476277079 |
- |
- |
DNA-3-methyladenine glycosylase, putative [Ricinus communis] |
| 17 |
Hb_000069_490 |
0.1483944095 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_016760_010 |
0.1499650463 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_003475_020 |
0.1506429079 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 20 |
Hb_004785_070 |
0.1516676643 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X2 [Jatropha curcas] |