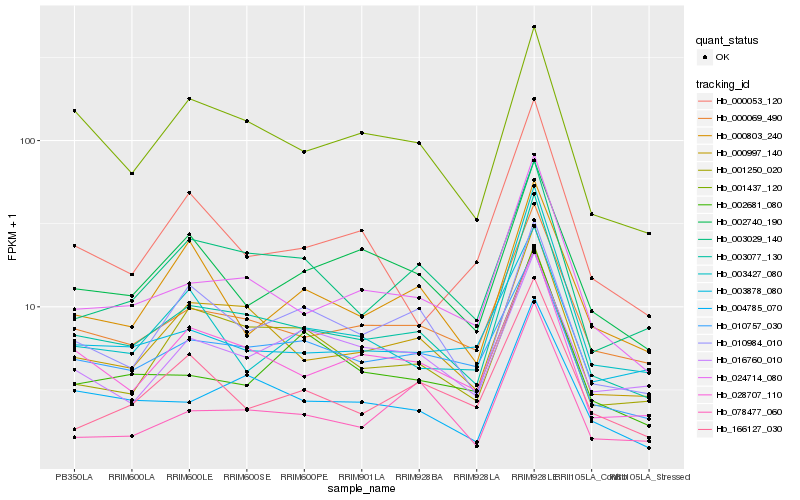
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003077_130 |
0.0 |
- |
- |
hypothetical protein JCGZ_22476 [Jatropha curcas] |
| 2 |
Hb_024714_080 |
0.0663617748 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000069_490 |
0.0915235794 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000997_140 |
0.0946753063 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_004785_070 |
0.0995166146 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_028707_110 |
0.1005695186 |
- |
- |
PREDICTED: uncharacterized protein LOC105645798 [Jatropha curcas] |
| 7 |
Hb_010757_030 |
0.1179842149 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_003029_140 |
0.1216452557 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 9 |
Hb_078477_060 |
0.1235512614 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 10 |
Hb_003878_080 |
0.124357438 |
- |
- |
PREDICTED: uncharacterized protein LOC105637045 [Jatropha curcas] |
| 11 |
Hb_003427_080 |
0.1250154449 |
- |
- |
PREDICTED: probable alanine--tRNA ligase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_166127_030 |
0.1270069113 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_002681_080 |
0.127470955 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 14 |
Hb_010984_010 |
0.1282324945 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_001437_120 |
0.1296120124 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Jatropha curcas] |
| 16 |
Hb_001250_020 |
0.1326182876 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000053_120 |
0.1353006078 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002740_190 |
0.1356372447 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000803_240 |
0.1400312942 |
- |
- |
hypothetical protein CICLE_v100143612mg, partial [Citrus clementina] |
| 20 |
Hb_016760_010 |
0.1403297522 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |